Circular DNA Amplification using TempliPhi™
Extracted from Nucleic Acid Sample Preparation for Downstream Analyses, GE Healthcare, 2007
illustra™ TempliPhi™ DNA Amplification Kits from GE Healthcare are novel products developed to exponentially amplify single- or double-stranded circular DNA templates by RCA, shown schematically in Figure 7.3 shown below. Amplified products can be used in downstream applications such as sequencing and molecular cloning. TempliPhi™ DNA Amplification Kit provides RCA reagents optimized for circular DNA amplification. The reaction utilizes modified random primers to efficiently prime synthesis by the extremely processive, high-fidelity Phi29 DNA polymerase in a single-step isothermal reaction.
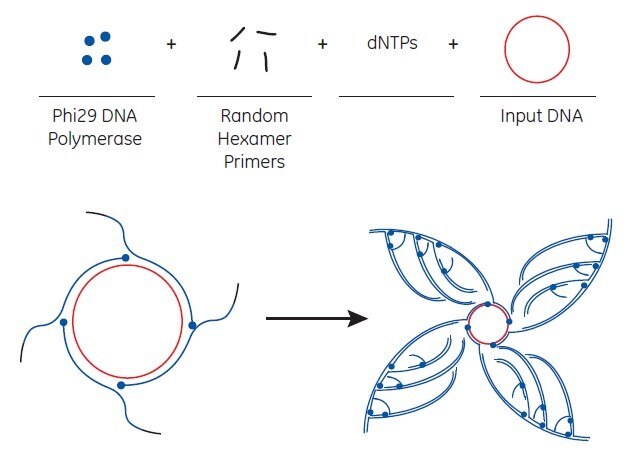
The starting material for TempliPhi™ amplification can be a variety of sources—a small number of bacterial cells containing a plasmid, an isolated plasmid, intact M13 phage, or any circular DNA (including ligation reactions) are efficiently amplified. Bacterial colonies can be picked from agar plates, added to a sample buffer, and heated to release the plasmid DNA from the cells. The polymerase is added directly to this material to initiate the amplification reaction. Alternatively, sub-microliter quantities of a saturated bacterial culture or a glycerol stock can be used. Depending on the source of starting material, amplification is completed in 4 to 18 hours at 30 °C with no need for thermal cycling. The product of the TempliPhi™ reaction is high-molecular-weight, double-stranded, tandem-repeated copies of the circular template. Note that when M13 clones are the source material, the TempliPhi™ product is double-stranded DNA that can be sequenced with both forward and reverse primers.
DNA amplified by the TempliPhi™ method can be used directly in many subsequent applications including cycle sequencing reactions, restriction enzyme analysis, DNA cloning procedures, and in vitro transcription reactions in place of supercoiled plasmid DNA. illustra TempliPhi™ DNA Amplification Kits are used to prepare DNA directly from circular DNA. illustra TempliPhi™ Large Construct DNA Amplification Kit is designed for large DNA vector (e.g., fosmid and BAC) preparation. illustra TemliPhi HT DNA Amplification Kit is designed for high-throughput use (over 1000 preps/wk). illustra TempliPhi™ Sequence Resolver Kit is designed to produce a different type of DNA that sequences more easily, solving the most common sequencing problems, and implements a simplified protocol with fewer steps and no overnight culturing.
A protocol for DNA amplification using TempliPhi™ Amplification Kits follows.
Materials
Sample buffer, reaction buffer, enzyme mix, and positive control DNA are provided with the product
Thermal cycler or water baths for incubations at 30 °C, 65 °C, and 95 °C
For single- or low-copy number BACs and fosmids: alkaline lysis reagents (see product instructions)
Advance preparation
Thaw sample buffer and reaction buffer on ice.
Protocol*
1. Mix sample buffer with template DNA†
2. Denature sample at 95 °C and cool
This step lyses bacterial cells or phage particles sufficiently to release the circular template into the liquid.
Avoid heating at higher temperatures or for longer than 3 min, which can release bacterial DNA that competes with the desired template during amplification.
3. Prepare enzyme master mix
Prepare the master mix for all the amplification reactions.
4. Transfer to cooled sample
Transfer enzyme master mix to the cooled sample.
5. Incubate at 30 °C
DNA is amplified during this step. The time will vary (4 to 18 h) with the amount of input DNA and desired amount of output DNA.
6. Heat-inactivate enzyme
Inactivate the enzyme by incubating briefly at 65 °C. Cool to 4 °C.
Inactivating the DNA polymerase activity prevents potential interference with subsequent cycle sequencing reactions. Inactivating the proofreading exonuclease activity prevents degradation of template DNA during storage. The heat inactivation step can be eliminated if the amplified DNA sample is used immediately.
* The protocol can be automated; Chapter 9.
† Alkaline lysis is recommended to denature cultures containing low-copy-number BACs and fosmids to reduce DNA fragmentation caused by thermal degradation; this is unnecessary for cultures containing plasmids or M13 phage.
如要继续阅读,请登录或创建帐户。
暂无帐户?