Whole Genome Amplification (WGA) Using GenomiPhi™
Extracted from Nucleic Acid Sample Preparation for Downstream Analyses, GE Healthcare, 2007
The GenomiPhi™ method of WGA from GE Healthcare uses Phi29 DNA polymerase, a highly processive enzyme with excellent strand-displacement activity, in combination with random-sequence hexamer primers (random hexamers) to amplify DNA in an isothermal process—a thermal cycler is not required. First, random hexamers anneal to multiple sites on the denatured linear DNA template. Next, Phi29 DNA polymerase initiates replication at these sites simultaneously. As synthesis proceeds, strand displacement of upstream replicated DNA generates new single-stranded DNA. Subsequent priming and strand displacement replication of this DNA produces large quantities of several-kilobase-long, double-stranded DNA suitable for genetic analyses that require high-molecular-weight product (Figure 1). Microgram quantities of high-molecular-weight DNA can be generated from as little as 10 ng of genomic DNA using this simple and robust procedure. DNA replication is extremely accurate due to the proofreading 3’–5’ exonuclease activity inherent in Phi29 DNA polymerase. Note that the GenomiPhi™ method does not preserve epigenetic methylation patterns.
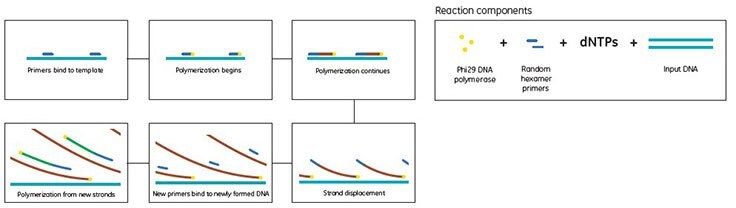
Figure 1.chematic showing the GenomiPhi™ method of genomic DNA amplification for WGA.
A protocol for DNA amplification using GenomiPhi™ Amplification Kits is provided below.
Materials
Sample buffer, reaction buffer, enzyme mix, and positive control DNA are provided with the product
Thermal cycler or water baths for incubations at 30°C, 65°C, and 95°C
Advance preparation
Thaw sample buffer and reaction buffer on ice.
Protocol*
1. Mix sample buffer with template DNA
2. Denature sample at 95 °C and cool†
Heating the DNA for longer than 3 min or at higher temperatures can damage the DNA.
3. Prepare enzyme master mix
Prepare the master mix for all the amplification reactions.
4. Transfer to cooled sample
Transfer enzyme master mix to the cooled sample.
5. Incubate at 30 °C
DNA is amplified during this step. The time will vary (1.5 to 18 h) with the amount of input DNA and desired amount of output DNA.
6. Heat-inactivate enzyme
Inactivate the enzyme by incubating briefly at 65 °C. Cool to 4 °C.
Heating is required to inactivate the exonuclease activity of the DNA polymerase which may otherwise begin to degrade the amplification product.
* The protocol can be automated; Chapter 9.
† As an alternative, genomic DNA template may be chemically denatured. See product instructions for details.
如要继续阅读,请登录或创建帐户。
暂无帐户?