Cloning Genes-of-Interest into a Plasmid Vector
Oxford Genetics
- Selecting the Cloning System and Plasmid Vector
- Plasmid Restriction Digestion
- Fragment Restriction Digestion
- Gel Excission
- Clean-up of Gel Fragments
- Annealing DNA Oligos for Ligation
- Adding 5’ Phosphates to DNA
- Dephosphorylating DNA
- Preparing DNA for Bunt-ended Cloning
- Ligating the DNA to yield a plasmid containing the Gene-of-Interest
- Site-Directed Mutagenesis
Selecting the Cloning System and Plasmid Vector
Genetic engineering is used in thousands of laboratories around the world. Given its importance it is remarkable that cloning strategies for many of the popular DNA components are not standardised. Contributing to this lack of standardization are the many options available for cloning kits and innovative technologies. It is not always necessary to use an IP- heavy kit, as traditional cloning by restriction enzyme digestion is made even easier with standardized plasmids from Oxford Genetics.
The aim at Oxford Genetics was to engineer a DNA plasmid system that could accommodate most of the functional DNA inserts that a researcher might require within a single plasmid. By optimising our starting vector, every DNA component of our system can be removed and exchanged for hundreds of other DNA sections that have been pre-designed and tested by us. This is the SnapFast™ concept. All of our constructs have been pre-screened for poor codon usage and conflicting restriction sites. Where possible, rare codons and restriction sites have been removed to enable efficient expression, and ensure that restriction sites do not limit the cloning of other SnapFast DNA sections.
We have one of the largest collections of plasmids available globally, providing multiple expression and cloning options for almost every insert we provide. Our product range includes:
- Largest peptide tag range anywhere (26)
- Total of 9 reporter genes in 5 configurations
- More than 20 signal peptides
- Over 40 promoters for mammalian, bacteria and yeast expression
- Total of 10 antibiotic and metabolic selection options
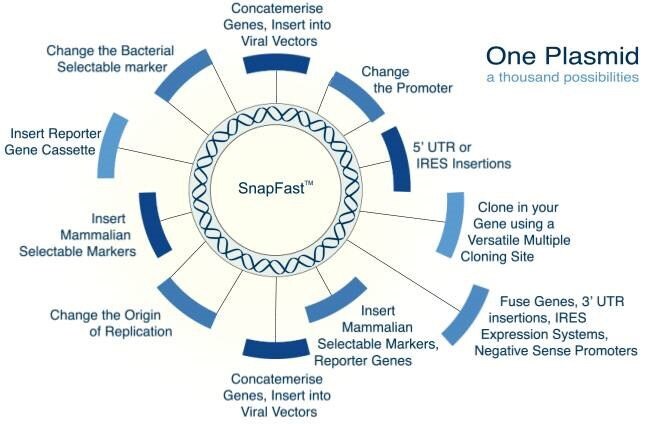
Benefits of the SnapFast system:
- All plasmids are compatible with standard cloning, LIC, InFusionHD, Gibson Assembly, Seamless Geneart.
- Over 1600 unique DNA sections are available and compatible.
- Easy, efficient, and simple engineering strategies.
- High success rates due to pre-designed compatiblity.
- No insert size constraints. Low copy origins for increased stability.
- Compatible with many pre-existing cloning vectors and a range of shuttle vectors to facilitate gene transfer.
- Easy cloning from the SnapFast vectors into a range of alternative systems, including viral vectors.
- Built in regulatory sequences and capabilities (e.g. gene concatamerisation, insulators and terminator sequences).
* Requires primer design using homology or TypeIIS sites
** Patents restrict the use of the cloning technology or products
^ Tools available online to help with design
LIC = Ligase Independent Cloning
Plasmid Restriction Digestion
A preparative digest is the cutting of DNA to prepare it for ligation with another piece of DNA, not simply to confirm the identity of the DNA. You should aim to set up the digest of both your fragment and vector at the same time. This will save you time. This protocol assumes a plasmid solution in Tris-EDTA (TE) or Elution buffer (EB) or nuclease free H2O that is 200-300 ng/µL and about 3-5 kb. Larger plasmids may require you to use more volume because at the same concentration there will be less plasmid copies.
In this particular example, the following could be done:
10-20 µL of plasmid (200-300 ng/μL, total amount 3-5 µg)
10 µL of 10X Restriction buffer
10 µL of 10X Bovine Serum Albumin (BSA, final concentration is usually 100 µg/mL) (Catalog No. A7906)
1.5-2 µL Restriction Enzyme 1 (10-20 units/µL)
1.5-2 µL Restriction Enzyme 2 (Optional)
Make up to 70 or 100µL total volume with TE (Catalog No. T9285) or nuclease free water (Catalog No. W4502)
Add the reagents above in a sterile 1.5 mL Eppendorf, first add the TE or water, then the plasmid/DNA, then the restriction buffer and BSA, and mix thoroughly. Finally, add the restriction enzyme(s). The restriction enzymes chosen depend on your goals and the plasmid map, but may include EcoR I, BamH I, or Hind III. Incubate the reaction at the right temperature (usually 37 °C, but always make sure that you are not dealing with an exception, like SwaI at 25 °C) and start your alarm clock (count up). Run the reaction for 40-60 minutes. Now you can de-phosphorylate the vector using alkaline phosphatase (CIP). You may wish to deactivate the restriction enzymes at this point, this is standard practice.
Run the digest on an agarose gel (with a very large well for loading a large sample), and inspect the results for a single band the size of your linearized plasmid. There are three possible outcomes from this digest:
- has not worked at all. There is no point in digesting longer because the reaction has failed. If the enzyme has worked on another plasmid before, perhaps re-precipitate the plasmid you are trying to cut, or do a new phenol extraction prior to precipitation and try again. Otherwise, check if the enzyme is still OK by using another plasmid.
- has worked but the vector is not completely cut after 40 minutes (visible as more than one band present at the approximate molecular weight you are expecting from the vector). You can estimate from the progress how much longer it will take and try another digestion.
- was complete digestion. You have a linear plasmid that is ready for use in a ligation procedure.
Tip: Restriction enzymes are very sensitive to temperature change so avoid putting them on ice. Ideally use a -20 °C cool box. Also, avoid holding them at the bottom of the tube with your fingers and try to work quickly whilst handling the enzyme stocks. They will last a long time if you look after them.
Fragment Restriction Digestion
This is a protocol for a preparative digest, which is the cutting of DNA to prepare it for ligation with another piece of DNA, not simply to confirm the identity of the DNA. Usually these pieces of DNA are circular plasmids but they could also be PCR fragments. You should aim to set up the digest of both your fragment and vector at the same time. This will save you time later.
This protocol assumes a plasmid solution in Tris-EDTA (TE) or Elution buffer (EB) or nuclease free H2O that is 200-300 ng/µL and about 3-5 kb. Larger plasmids may require you to use more volume because at the same concentration there will be less plasmid copies.
In this particular example, the following could be done:
10-20 µL of plasmid (200-300 ng/μL, total amount 3-5 µg)
7.5 µL of 10X Restriction buffer
7.5 µL of 10X Bovine Serum Albumin (BSA, final concentration is usually 100 µg/mL) (Catalog No. A7906)
1.5-2 µL of Restriction enzyme 1(10-20 u/µL)
1.5-2 µL Restriction Enzyme 2 (Optional)
Make up to 70 µL total with TE (Catalog No. T9285) or nuclease free water (Catalog No. W4502)
DNA Fragment Restriction Digestion Protocol
Add the reagents above in a sterile 1.5 mL Eppendorf, first add the TE or water, then the plasmid/DNA, then the restriction buffer and BSA, and mix thoroughly. Finally, add the restriction enzyme(s). Incubate the reaction at the right temperature (usually 37 °C, but always make sure that you are not dealing with an exception, like SwaI at 25 °C) and start your alarm clock (count up). Run the reaction for 40-60 minutes. Run the digest on an agarose gel (with a very large well for loading a large sample), and inspect the results for a single band the size of your insert.
Gel Excission of DNA Fragments
To excise the band you will need a clean scalpel or razor blade. An important consideration is the amount of agarose you take with DNA. The higher the ratio is between the quantity of DNA to agarose the less contaminants you will get carrying through to the ligation.
The standard method used to extract DNA from agarose gels is by visualization of ethidium bromide (Catalog No. H5041) stain with UV light. This is not ideal because the UV will damage the DNA and can cause mutations and can reduce the ligation efficiency. An alternative is to use a Clare Chemical Dark Reader Trans-illuminator but it can sometimes be difficult to see low quantity bands. If you have a signficant amount of DNA these systems are ideal. Alternatively, you can do a blind excision by running a small quantity of your cut DNA in the lane adjacent to your large well, and use its position to excise a large quantity of DNA in the next well. Be very careful with this technique as some bands can be quite small lengthways/top-to-bottom in the gel and can be missed if you are not accurate. After excision you can look at the remaining gel to see if you got the band, do it quickly because if you didn't get the DNA you don’t want to damage it with the UV.
Be careful of DNA fragments that are close together. The key of this technique is to separate the vector backbone from the fragment to prevent contamination. Just because you cannot see something when you are excising doesn’t mean it isn’t there, you will only see the peak of a Gaussian distribution of DNA and whilst the bands may look separate they may not be and you may get some DNA from bands that are close by accident.
Tip: This is a relatively dangerous procedure as far as molecular biology goes. UV light, carcinogenic ethidium bromide, and a scalpel are a recipe for disaster. Please abide by all local safety rules and regulations and consult with the local safety officer if you are unsure about the necessary precautions to take.
Tip: Consider choosing a safer chemical for visualizing DNA. Both Nancy 520 (Catalog No. 01494) and Syber Safe (Catalog Nos. S9305 and S9430) have been shown to be less mutagenic than ethidium bromide.
Clean Up of Gel Fragments
For gel extracting a DNA fragment from an agarose gel, it usually isn’t worth using anything other than a prepared kit, like the GenElute™ Gel Extraction Kit, Catalog No. NA1111. They are relatively cheap and highly effective. Other methods exist that don’t involve a kit (like dialysis tubing and squashing the gel fragment between parafilm to squeeze out the DNA in solution) but often using these methods result in high levels of contaminants and low DNA yields.
DNA gel extraction kits usually consist of either beads or spin columns. The sizes of DNA fragments that each kit can accommodate can vary, so check that the kit will extract the size range of DNA you are working with. Most kits cover from 100bp – 5 Kb but some have lower or higher capacities.
The principle of both spin column and bead kits is to bind the DNA to something (usually a silica resin) and then wash away the contaminants, either by pelleting the beads, or washing the column. The final elution step releases the DNA from the resin/beads/column to provide you with a clean DNA fragment. However, even the best extraction kit can still carry though some contaminants and, for this reason, using large volumes of a solution that has been produced by gel extraction in a ligation should be avoided. Sometimes ‘adding less is more’ when it comes to adding your fragment to your ligation. Try to avoid making a ligation consist of more than 30% volume of liquid isolated by gel extraction.
Gel Extraction Kit Variability
We have tested six different DNA gel extraction kits in our laboratories, head-to-head, because the process is so essential to our work. We have found that whilst some kits constantly provide high yields and pure DNA, others sometimes provide good yields but sometimes fail, and others consistently under-perform.
If you gel extract a fragment from a plasmid, you should probably expect the DNA concentration numbers (if using a spectrophotometer) to be low, because you will have started with a large plasmid (most likely around 5kb), then cut out a much smaller sub-fraction (perhaps 1kb), and the kits will never purify 100% back of the DNA from the gel. So a yield of 20ng/ul in 35ul total volume for 1Kb fragment isolated from 5ug of plamsid DNA (5Kb) would not be too bad (70% of the DNA has been recovered).
We frequently find that often the level of contaminants, or DNA prep quality, is a more important determinant of cloning success, rather than the DNA yield itself. We have also consistently observed that sometimes adding more fragment to a ligation reduces the cloning success compared to the same reaction with slightly less. Although, this could simply be because the extra DNA ends are titrating the ligase away from the vector ends, adding too much gel extracted material may be contributing because of low levels of contaminants in the prep from the gel.
Annealing DNA Oligos for Ligation
The basic concept of annealing oligos is to heat two oligonucleotides up such that they denature, then follow this by a period of cooling to allow the two oligos to base pair together. For custom oligos, see OLIGO). This process is often used to prepare short DNA sections for:
- Creating shRNA DNA regions for ligation
- Creating microRNA DNA regions for ligation
- Adding a linker to remove or add a restriction site.
- Studies requiring small double stranded DNA regions, such as DNA protein binding assays.
The cost of purchasing lots of phosphorylated oligos can be prohibitive. For this reason many groups simply ligate oligos into vectors that have not been de-phosphorylated. This is particularly effective when the oligos are to be ligated into a vector that has been cut with two separate restriction enzymes that have incompatible ends (preventing the vector from closing on itself). The background from the ligation may still be high but the oligos are often in significant excess in oligo ligations and so the reaction should still work.
It is also possible to phosphorylate the oligos first using polynucleotide kinase (PNK), but cleaning up the oligos is difficult because they are likely not to bind a DNA clean up column because of their short length. However, PNK reactions are performed in ligase buffer so this may not be necessary, but the PNK must be heat inactivated before the oligo is put into the ligation otherwise the PNK will phosphorylate your vector.
Phosphoramidite chemistry, by which oligos are generally produced, can normally be relied on to produce 40-50 bases without too many mistakes, however, using this technique to produce oligos of 100 bases or longer often leads to mutations. We have frequently found that it is better to divide a long oligo into two shorter oligos with 10-15 base pairs of overlap in the middle on the opposing oligo to allow them to join together. The frequency of mutations you will get following sequencing should be much lower.

Consider the cooling step. We have tried cooling the oligos in a water bath simply by heating it to 95 degrees and then turning it off to cool down, or by heating them to 100 degrees in a beaker on tripod (old school) and then placing the beaker in iced water (and oligos in a tube within the beaker still) to cool. We have also tried placing the oligos in PCR machine set with 5 degree cooling increments every 30 seconds. The reality is that the method does not make a huge amount difference. Leaving them for too long at high temperature could lead to hydrolysis, hence we avoid just turning the water bath off. We generally heat the oligos to 95 degrees in a water bath in a beaker with the tube of oligos in an eppendorf on a float. Then place this in iced water for 10 minutes. This seems to work well for us, but the other methods may also work for you.
Designing oligos with overhangs that reconstitute the sites into which the oligo is to be inserted can be difficult. This diagram below provides an example of what the oligos may look like, in this example the oligos are to be ligated NcoI and XbaI restriction sites.

Annealing DNA Oligos Protocol
Tip: Oligo and primer stocks are often resuspended at 100 µM (100 picomoles/ul) concentrations.
- Add 10 µL of stock oligos (assuming you have two to anneal) to 25 µL nuclease free water (Catalog No. W4502) into a 1.5 mL microtube.
- Add 5 µL restriction digest buffer (such as 100mM NaCl (Catalog No. S3014), 50 mM Tris-HCl (Catalog No. 93362), 10 mM MgCl2 (Catalog No. M0250), 1 mM Dithiothreitol (Catalog No. D0632), pH 7.9. The presence of the buffer helps to keep the pH correct for DNA stability and the salt should promote annealing. The oligo concentration is now at 20 picomoles/ul.
- Float the tube in a beaker in water pre-heated to 95 degrees for 5 minutes.
- Place the beaker into an ice box that is filled with ice and water to allow the beaker to cool for 10 minutes.
- Once the water in the beaker has cooled dilute the oligos 10-fold and 100-fold in water and add 1 µL of this to a standard 20 µL ligation reaction. The temptation is to add more but this will just titrate the ligase away from the ends of your vector onto the extra oligos, which can reduce the ligation efficiency. The final concentration of oligos in the ligation would be approximately 2 or 0.2 picomoles/µL. This will still be a significant excess of oligos to vector.
Adding 5' Phosphates to DNA
T4 DNA ligase requires a 5’ phosphate on one of the DNA molecules to be ligated in order to join DNA, for this reason it is often necessary to phosphorylate DNA molecule prior to adding it to ligation, for example when blunt cloning a PCR product.
DNA Phosphorylation Protocol
Nuclease free water (Catalog No. W4502): 4.5 ul
PCR product or other DNA (cleaned-up): 4 ul (if your PCR product has 5' recessed or blunt ends, heat it to 70 °C for 5 minutes and cool on ice before adding it to the reaction)
10 x T4 DNA Ligase buffer: 1 ul (ligase buffer is used because it contains ATP)
T4 Polynucleotide kinase: 0.5 ul
Incubate the kinase reaction(s) for 30 minutes at 37 °C. The phosphorylated PCR product can then be used directly in a ligation reaction without clean-up (for example when performing site-directed mutagenesis). If the PCR product is to be ligated into a de-phosphorylated vector, heat-inactivating the PNK enzyme may be a good idea to prevent it from phosphorylating the backbone vector and leading to high background. This is achieved by incubating the reaction(s) for 20 minutes at 65 °C.
Dephosphorylating DNA
The purpose of de-phosphorylating the vector is to prevent it from ligating back on itself during the ligation step by removing the 5' phosphate groups that are required by DNA ligase to join the phosphodiester DNA backbone together. Various alkaline phosphatases exist, including Calf Intestinal Phosphatase (CIP), Shrimp and Antarctic phosphatases. CIP (Catalog No. P4978) is the most commonly but it is difficult to heat inactivate. The temperature and buffers of the different enzymes can be different, refer to the manufacturer’s instructions.
DNA Dephosphorylation protocol for dephosphorylating DNA in a restriction digest reaction
50-100 µL of DNA (5µg) in a restriction digest reaction/solution
1-2 µL of CIP enzyme (1unit/µL)
- Add 1-2 µL of CIP to your restriction digest. CIP is stable and active in most restriction digestion buffers.
- Incubate the sample for 30-60 minutes at 37 °C.
DNA Dephosphorylation protocol for dephosphorylating DNA in TE or H2O
20-40 µL of DNA (5µg) in TE (Catalog No. T9285) or nuclease H2O (Catalog No. W4502)
5 µL of 10xCIP buffer
1-2 µL of CIP enzyme (1unit/µL)
Make up to 50 µL
- In a sterile 1.5 mL eppendorf, add the DNA, then the CIP buffer and then the 1-2 µL of CIP.
- Mix thoroughly with a pipette tip and incubate for 30-60 minutes at 37 °C.
Tip 1: Make sure your fragment you are going to ligate into the dephosphorylated vector possesses 5’phosphate groups. Standard oligos/primers and PCR products are generally not phosphorylated and must be treated with T4 polynucleotide kinase (see phosphorylating 5’ ends). It is usually easier to add restriction sites to the ends of a PCR product (plus a few extra base pairs at the ends), rather than phosphorylating a fragment.
Tip 2: CIP is stored in a glycerol buffer for stability, but this means it sinks to the bottom of aqueous solutions. When adding the CIP, watch it drop into the DNA mixture by doing it with the tube held up in front of you, and then ensure it is properly re-suspended before incubation.
DNA Blunt Cloning Protocols
Sometimes it is necessary to make the ends of a DNA molecule blunt, for instance:
- When making a DNA library
- Sheared DNA with ragged ends that need to be cloned into a vector
In a situation when choosing compatible restriction sites is impossible requiring the vector and insert, or both, to be blunted before ligation (may be a last resort). There are two options, either use a DNA polymerase (like Klenow or T4) or Mung bean nuclease. Klenow and T4 DNA polymerases both fill-in 5’ overhangs and chew back 3’ overhangs. If you need to fill in a 5’ overhang either enzyme will be fine but if you need to remove a 3’ overhang then T4 may be a better choice as it has a stronger 3’ to 5’ exonuclease activity. Mung bean nuclease chews back both 5’ and 3’ overhangs.
Klenow Blunting Protocol
- DNA should be dissolved in 1x restriction digest buffer or T4 DNA Ligase Reaction Buffer supplemented with 33 μM each dNTP [final concentration].
- Add 1 unit of Klenow per microgram of DNA and incubate for 15 minutes at 25 °C.
- Stop the reaction by adding EDTA to a final concentration of 10mM and then heating at 75 °C for 20 minutes.
T4 Blunting Method
- DNA should be dissolved in 1x restriction digest buffer & supplemented with 100 µM dNTPs [final].
- Add 1 unit T4 DNA Polymerase per microgram DNA and incubate for 15 minutes at 12 °C.
- Stop the reaction by adding EDTA to a final concentration of 10 mM and heating to 75 °C for 20 minutes.
Mung Bean Nuclease Blunting Method
- Suspend DNA (0.1 μg/μL) in 1X Mung Bean Nuclease Buffer or a restriction enzyme buffer
- Add 1.0 unit of Mung Bean Nuclease per μg DNA.
- Incubate at 30 °C for 30 minutes.
DO NOT attempt to heat inactivate Mung bean nuclease because single stranded regions of DNA may appear before the enzyme is inactivated resulting in unintended degradation. Inactivate the enzyme by spin column purification or by phenol/chloroform extraction and ethanol precipitation.
DNA Ligation Protocol
During the cloning process, all roads lead to the ligation, and so all of the steps that precede it can affect its efficiency significantly. It is important to read the tips and notes in each section before the ligation reaction.
Ligation reactions are set up along with controls. Usually you have two controls which are:
- The vector alone without ligase (controls for uncut vector)
- The vector with ligase (controls for insufficient dephosphorylation when used in conjuction with control 1)
- The real ligation which is vector + fragment + ligase.
If you got colonies on control 1 then your restriction digest of your vector did not work because you got colonies even without ligase. If you got colonies on control 2 but not 1 then your alkaline phosphatase treatment didn’t work. This is because your digest worked well (no colonies on control 1) but the ligase was able to re-circularise the plasmid because of the 5’ phosphates were not removed during dephosphorylation. If you got colonies on 3 and none (or just less) on 1 and 2 then you should congratulate yourself, pick some colonies and go home for a cup of tea to celebrate.
Reaction conditions can vary between labs. Best results are achieved at 16 °C overnight but this means adding an extra day to the whole process (making it a 4 day cloning cycle). Performing a ligation at room temperature for 1-2 hours usually gives good results and shortens the cloning to 3 days for a complete cycle. This is provided you don’t mind a long first day.
The two most difficult types of ligations are ligating PCR products and blunt ligations. You should expect these to be less efficient than standard cloning of a fragment from one vector to another.
A typical reaction could be set up as follows:
- Start setting up the reaction by adding the components that are shared for all reactions i.e. add the water then the buffer and then the vector to each of the tubes.
- Then add the fragment to the ligation tube.
- Finally, add the ligase to the ligation tube and the correct control tube. Do not vortex the reaction as DNA ligase is shear sensitive. Instead mix with the tip you added the ligase with by stirring well and/or flicking the tube gently repeatedly with your finger.
- Incubate for either 1-2 hours at room temperature, or 16 °C overnight. Some people heat inactivate the ligation at 65 °C for 20 minutes but this is not strictly necessary.
- Once ligation is completed, use the plasmid to transform bacteria so you can either express protein directly, or grow many copies of the plasmid DNA for further use.
Tip 1: The BSA in the ligase buffer can precipitate out on freeze thawing, visible as a white precipitate at the bottom of the buffer. Re-suspend by vortexing and temporarily warming it between your fingers or at 37 °C.
Tip 2: The ligase buffer contains ATP, which degrades after multiple freeze thaw cycles. Keep the buffers stocks in small aliquots and throw away after >3 cycles.
Tip 3: Heating your ligation (before adding enzyme) to 37 °C for a few minutes to open up the sticky ends, or help to linearise transiently re-cirularised vectors when using single restriction site ligations. Cool the ligation back to room temperature before adding the enzyme to avoid damaging the ligase when you add it. We dont do this, but we have heard it can help from others who do.
Tip 4: Treating PCR products with Proteinase K before ligation is reported to help with the ligation. Gel extracting the PCR product would make this unnecessary, as would using a PCR clean up kit to purify it.
Tip 5: Avoid exposing your DNA to UV when extracting it from a gel. This can reduce ligation efficiency dramatically in some cases.
Tip 6: Avoid using more than 20-30% of the total ligation volume (normally the total volume is 20 μL) of gel-purified material. If you do, you may have too much salt and other contaminants in your reaction and the ligation efficiency may be reduced.
Tip 7: The amount of vector should be barely visible if you were to run it on an agarose gel, so about 20-30 nanograms. Fragments should be more abundant, but not more than 5-10-fold concentrated compared to the vector. We normally use a 1:2 or 1:3 vector to fragment ratio.
Calculating the insert to vector ratio
The insert to vector molar ratio can have a significant effect on the outcome of a ligation and subsequent transformation steps. Molar ratios can vary from a 1:1 insert to vector to a ratio of 10:1. It may be necessary to try several ratios in parallel for best results. The calculation below will tell you the amount of fragment to us relation to the vector at a ratio of 6:1 (fragment:vector). Change the 3 to anything to work out alternative concentrations.

Site Directed Mutagenesis Protocol
Site Directed Mutagenesis (SDM) is a useful technique for introducing a specific mutation into a plasmid at a specific site. The mutation can be a substitution, insertion or deletion. There are many applications for SDM, for instance to assess the function of certain amino acids in an enzyme, to investigate the effect of altering bases in a promoter or removing restriction sites from a plasmid.
Here we describe a PCR based site directed mutagenesis method. The basic principle is to design a pair of PCR primers back to back, so that the entire plasmid is amplified by PCR. One of these primers incorporates the desired mutation. The PCR creates a linear product whose ends can then be joined together (after phosphorylation) with DNA ligase. The circularised vector is transformed into E. coli.
Step 1: Mutagenesis Process and Primer Design
Design a pair of primers incorporating your desired mutation into the 5' end of one of them. Design them so that the complementary region has a Tm of around 60 °C.
Example:
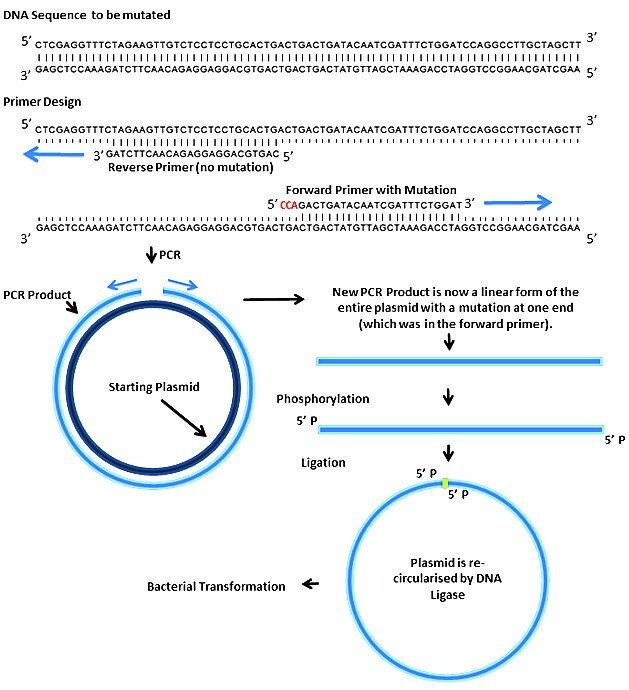
Note that only the forward primer contains the mutation, so you could easily make a series of different mutations by keeping the reverse primer the same but using different forward primers. The above example is a 3 bp substitution but an insertion can be made in the same way. If you require a very large substitution or insertion, then mutant bases may be introduced at the 5' end of both primers.
Deletions of any size can be made by spacing the forward and reverse primers apart on the template.
PCR primers normally come without a phosphate group on their 5' termini due to the way they are synthesised. This means that the ends of a PCR product cannot simply be ligated together, they must be phosphorylated first. There are two main options for this: 1) you could order the primers with a phosphate already added to the 5' end, or 2) you could phosphorylate the PCR product using polynucleotide kinase (PNK). Phosphorylated primers are a good idea if you are only doing a few SDMs and don't have any PNK in the freezer (plus it cuts out a step). Using PNK is more cost effective if you're doing a lot of SDMs (probably 10 or more).
Step 2: PCR
It is important to use a proofreading polymerase to avoid introducing any other mutations. That said, if you were still worried about introducing mutations you could sub-clone the mutated fragment back into the same backbone after the mutagenesis.
Because the method is based on PCR, it tends to work better on smaller plasmids. It is best to follow the instructions that come with the polymerase, but set up a PCR similar to this example:
35.5 μL water
5 μL 10x polymerase buffer
1.5 μL Forward primer (0. 3μM final)
1.5 μL Reverse primer (0.3 μM final)
5ul dNTPs (200 μM final)
1ul Template DNA (a 1 in 100 dilution of a mini-prep)
0.5 μL polymerase
Set the reaction up on ice, and transfer the tube to a pre-heated PCR block straight after mixing the reaction.
PCR program:
- 98 °C 60 seconds
- 98 °C 8 seconds
- 55-65 °C 20 seconds
- 72 °C (the elongation time depends on the plasmid size and the type of polymerase used) repeat/cycle steps 2-4 a further 27-30 more times
572 °C 5 minutes
Hold at room temperature.
Step 3: Clean up the PCR product
Run the entire reaction on an agarose gel. Excise the band and clean up the DNA using a gel extraction kit, eluting it in 30 μL.
Step 4: Phosphorylate the 5' termini
*This step can be omitted if you used phosphorylated oligos in the PCR.
4.5 μL Nuclease free water (Catalog No. W4502)
PCR product
1 μL 10x T4 DNA Ligase buffer*
0.5 μL T4 Polynucleotide kinase
Incubate at 37 °C for 40 mins.
*Ligase buffer is used because it already contains ATP and PNK is active in it.
Step 5: Ligate the DNA ends
Set the ligation reaction up on ice.
6.7 μL Nuclease free water (Catalog No. W4502)
2 μL PNK Reaction (from step 5)
0.8 μL 10x T4 DNA Ligase buffer
0.5 μL T4 DNA Ligase
Incubate at 16 °C overnight or at room temperature for 2 hours.
Step 6: Transform into competent E. coli
Step 7: Confirm the mutation by DNA sequencing

如要继续阅读,请登录或创建帐户。
暂无帐户?