Histone Modification and Chromatin Remodeling | Epigenetics
Savita Bagga, Ph.D.
Histone Modification and Gene Expression
Gene expression is governed by complex mechanisms including transcription factor binding to DNA and coordinated changes in chromatin structure. The primary protein components of chromatin are the histones, which are assembled along with DNA into larger complexes known as nucleosomes. Each nucleosome contains two copies of the core histones, H2A, H2B, H3, and H4, each of which has an accessible amino terminal tail with a high proportion of lysines and arginines. Modifications of histone proteins constitute an important mechanism of gene regulation.
Histone modifications are an early indicator of epigenetic regulation, and one way to study this phenomenon is via chromatin immunoprecipitation (ChIP). ChIP monitors DNA-protein interactions and allows the chromatin structure surrounding a specific DNA sequence to be analyzed. This technique, used with histone modificationspecific antibodies, identifies and quantifies those genomic regions containing the targeted histone modifications. The protocol starts with cells, and uses a cross linking agent to chemically link the DNA and it's interacting proteins. The resulting DNA is isolated, sheared, and precipitated using a protein-specific antibody (e.g. acetylated histone). The cross links are reversed or broken, and the precipitated DNA, now enriched for sequences that interact with the protein of interest, is examined to determine which genomic regions are present. Detection can be via PCR when looking for a few genes, or can be done using microarrays (ChIP-chip) or parallel (deep) sequencing (ChIP-seq).
Chromatin Immunoprecipitation (ChIP) Kit (CHP1)
We combine speed and convenience with performance in our Imprint Chromatin Immunoprecipitation Kit (CHP1). The Imprint ChIP Kit provides a complete solution for chromatin immunoprecipitation including columns and reagents for DNA purification. Our CHP1 Kit is designed for abundant targets, such as histone modifications, and is available in a plate format. The kit affords a high-throughput system, ideal for researchers screening multiple samples for histone modifications.
Features and Benefits
- Fast – Total protocol time of less than 6 hours, making the Imprint ChIP kit the fastest on the market
- Sensitive – As few as 10,000 cells required for each ChIP sample
- Convenient – Fewest steps of any available ChIP protocol
- Flexible – Protocols for cells or tissue, and convenient strip-well format for high-throughput applications
- Complete – Includes columns and reagents for DNA purification as well as an integrated protocol for amplification with the GenomePlex® technology
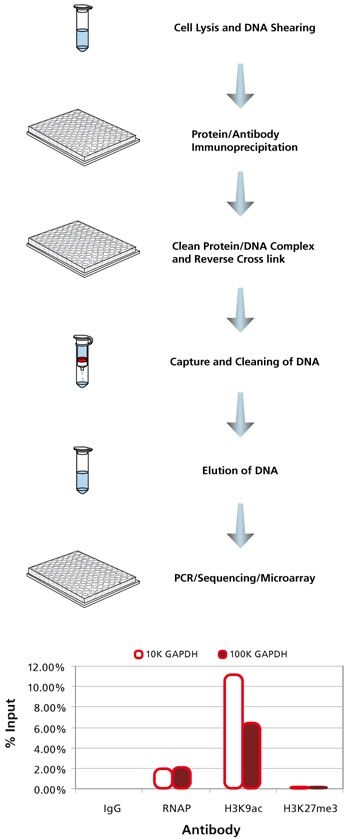
One day is sufficient to acquire ChIP enriched DNA using our Imprint CHP1 protocol. ChIP with antibodies against mouse IgG, H3K9ac, RNAP, and H3K27me3 was completed on 10,000 and 100,000 SW480 cells per reaction. A fraction (1%) of the sonicated chromatin was set aside as “input” DNA before the antibody affinity manipulations. DNA was purified following cross link reversal. The resulting enriched DNA was probed for GAPDH using specific primers by qPCR. Percent input was calculated by 100 x 2^(Ct adjusted Input – Ct Enriched). Input DNA Ct was adjusted from 1% to 100% equivalent by subtracting 6.644 Cts or Log2 100.
Chromatin Immunoprecipitation (ChIP) Validated Antibodies
Chromatin immunoprecipitation (ChIP) is a powerful tool for identifying proteins, including histone proteins and non-histone proteins, associated with specific regions of the genome by using antibodies that recognize a target protein or a protein modification. Our ChIP kits allow for a rapid and high-throughput approach, as well as high levels of sensitivity, to the study of DNA-protein interactions. Equally important to the ChIP kit is the selection of an antibody. Successful chromatin immunoprecipitation requires an effective, specific, and high quality antibody. Furthermore, not all antibodies work in ChIP, as the antigen epitope may be blocked or altered once cross-linked. We have a growing collection of validated antibodies. Each lot of these antibodies is validated for chromatin immunoprecipitation.
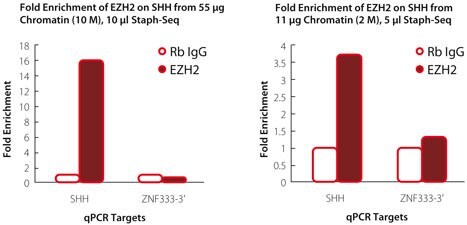
ChIPs were performed with 2 µL of EZH2 Ab (Diagenode, pAb39) and 1 µL of Rabbit IgG (Product No. I5006) with chromatin from DU145 cells as indicated (10 million cells on left panel and 2 million on right panel). 2 µL of ChIP'ed DNA (out of 30 µL eluate) was analyzed by qPCR using primers for the sonic hedgehog (SHH) gene promoter, a target for EZH2 containing polycomb repression complex, and a non-target ZNF333 gene (ZNF333-3').
Whole Genome Amplification Technology (WGA)
The GenomePlex® Whole Genome Amplification (WGA) family of products provide a robust and accurate method of amplifying nanogram quantities of starting material into microgram yields with minimal allele drop out. It utilizes a proprietary amplification technology based upon random fragmentation of genomic DNA and conversion of the resulting small fragments to PCR-amplifiable OmniPlex® library molecules flanked by universal priming sites. The OmniPlex library is then PCR amplified using universal oligonucleotide primers and a limited number of cycles.
The GenomePlex Whole Genome Amplification kits have been very successfully applied to ChIP DNA amplification and is the method of choice for generating more copies of a fragmented DNA sample. WGA has been used in a variety of applications including amplifying DNA from formalin-fixed paraffin-embedded (FFPE) tumor tissue, genotyping single-nucleotide polymorphisms (SNPs), and amplifying single copies of chromosomes. It is suitable for use with purified genomic DNA from a variety of sources. GenomePlex WGA uses nanogram quantities of starting genomic DNA, which after PCR yields 5–10 µg of WGA product. After purification, the WGA product can be analyzed in a manner similar to any genomic or chromosomal DNA sample. In addition to ChIP DNA, WGA can be used with purified genomic DNA from a variety of sources including blood cards, whole blood, buccal swabs, soil, plant, and FFPE tissues.
Whole Genome Amplification Sequencing Technology (SEQX)
The SeqPlex DNA Amplification Kit for whole genome amplification (WGA) is designed to facilitate next-generation sequencing (NGS) from extremely small quantities or from degraded/highly fragmented DNA. The yields from chromatin immunoprecipitation (ChIP) or formalin-fixed paraffin-embedded tissue samples (FFPE) are often less than required for successful NGS library preparation. The SeqPlex kit allows the user to pre-amplify these and other small quantity/highly fragmented DNA samples for input into a NGS workflow.
This kit is an extension of the WGA product line and has been developed to integrate into the Illumina, Solid, or 454 sequencing workflows.
Whole Transcriptome Amplification Technology (WTA)
The TransPlex® Whole Transcriptome Amplification (WTA1 and WTA2) kits provide an accurate, fast, and simple method of amplifying total RNA from a variety of sources including blood, fixed and frozen tissue, cell culture, FACS-sorted cells, plants, and microorganisms. The TransPlex WTA technology accurately amplifies total RNA by the utilization of a unique blend of quasi-random primers to ensure accurate transcriptome coverage and rapid amplification of total RNA. The resulting cDNA is suitable for qPCR, microarray, and traditional cloning.
Histone Modifications
Eukaryotic DNA is highly organized and packaged into the nucleus as chromatin. The primary protein components of chromatin are the core histones H2A, H2B, H3, and H4, which together with DNA form chromatin. In eukaryotes, the packaging of DNA in chromatin regulates DNA metabolic processes such as transcription, replication, and DNA repair. Chromatin structure and function can be affected by various post-translational modifications of the amino-terminal tails of nucleosomal histones. Acetylation results in the loosening of chromatin and lends itself to replication and transcription. The level of acetylation is related to transcription activity. Acetylation induces an open chromatin confirmation that allows the transcription machinery access to promoters. Histone deacetylases (HDACs) and histone acetyltransferases (HATs) are enzymes that influence transcription by selectively deacetylating or acetylating the e-amino groups of lysines located near the amino termini of core histone proteins. HDACs are also involved in the reversible acetylation of non-histone proteins. HATs and HDACs have major roles in the control of cell fate and their misregulation is involved in diseases like cancer and chronic asthma.
Histone Deacetylases
Histone deacetylases (HDACs) are enzymes that remove acetyl groups from lysines of histones and a number of other regulatory and structural proteins. They play critical roles in chromatin remodeling and are involved in transcription regulation, cell-cycle progression, cell survival, and differentiation. Eighteen HDACs have been identified and divided into three distinct enzyme classes.
Histone Acetyltransferases (HAT)
Histone acetyltransferases (HAT) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyl lysine. Histone acetyltransferases (HATs) generally act as transcriptional coactivators.
Histone Methylation
In the cell nucleus, DNA is wound around groups of histone proteins. Changes in the methylation patterns of these histone proteins impacts gene expression levels. Methylated histones bind DNA more tightly, which inhibits transcription while unmethylated histones lead to increased gene expression. Histone methylation can be broadly grouped into two families, lysine methylation and arginine methylation.
如要继续阅读,请登录或创建帐户。
暂无帐户?