Precision Genetic Modifications
Project Types
Our scientists’ years of experience and access to multiple advanced gene editing technologies allow us to offer a cell engineering service with unparalleled success. We can engineer a variety of modifications in your cell line of choice to address your scientific questions.
In most projects, desired modifications are introduced with nuclease cleavage to produce a lesion at your locus of interest. Projects can be broadly split into those that exploit errors introduced via the non-homologous end joining (NHEJ) repair pathway, and those that depend upon the homology-directed repair (HDR) pathway, which utilizes the homologous recombination machinery to insert donor sequence into a specific position at the locus of interest. Examples of these projects include:

- Knockout
- Traditional knockout: NHEJ repairs the lesion in an error-prone fashion, producing a frameshift mutation. Frameshifts typically cause a truncated mRNA because of the introduction of a new, premature stop codon. These truncated transcripts can be degraded by nonsense-mediated decay, resulting in no peptide; when truncated peptides are produced they are typically non-functional.
- Knockout by knockin: A reporter gene (or other gene) with its own promoter is inserted into an existing gene using a donor molecule and HDR. The insertion of non-native sequence interrupts the target locus, resulting in similar consequences for the gene of interest as a traditional knockout.

- Knockin (targeted integration)
- Fusion proteins: A reporter gene, tag, or other sequence is added to an existing coding sequence using a donor molecule and HDR. The donor molecule is designed such that the reporter or tag is in frame with the targeted gene to produce a single polypeptide and ideally, a functional protein.
- Promoter reporters: A reporter gene is inserted downstream of an endogenous promoter using a donor molecule and HDR. The reporter is inserted in place of the endogenous coding sequence in at least one allele, thus acting as a readout of endogenous promoter activity.
- Expression of a gene from a “safe” locus: A gene of interest is inserted into a defined locus considered both safe and transcriptionally active using a donor molecule and HDR. Examples of these sites include AAVS1 in human cells or ROSA26 in murine cells, which allow for transgenic expression without the risks inherent in random integration.
- Gene modification: A donor sequence harboring a particular mutation, such as a SNP, is inserted as in a typical knockin/targeted integration. This allows for gene editing at the single-nucleotide level, such as correction of disease mutations in patient-derived iPS cells or introduction of SNPs into a model cell line.
In addition to the services described above, where site-specific nucleases are used for genome modification, we also offer random integration services. We can achieve stable expression of a transgene or shRNA by stable transfection or lentiviral transduction as dictated by your needs or the cell line.
Depending on your needs, we can deliver cells to you either as individual clones produced from single cells or as a modified pool of cells.
Cell Engineering Technologies
CRISPR/Cas9
We offer the newest in CRISPR gene editing technologies. Cell Design Studio uses both codon-optimized Cas9 (wild-type nuclease activity) and Cas9-D10A nicking nuclease for use as a paired nickase. We can use pre-validated gRNAs or produce bioinformatically optimized custom gRNAs to meet your needs. We also offer integrated/stable Cas9 lines for high-throughput knockout screens. Learn more about CRISPR/Cas9 editing here.
Zinc Finger Nucleases
As the first company to commercially offer genome editing technologies, scientists are the world’s experts in zinc finger nuclease design and utilization. Cell Design Studio uses CompoZr® ZFN technology as a mature, affordable genome editing tool. We can also extend commercial licensing terms for ZFN-derived lines. Learn more about ZFNs here.
RNAi
We also produce shRNA-expressing lines for RNAi-based pooled screening projects. MISSION® shRNA offers the advantage of long-term, stable gene knockdown using established technologies. Learn more about shRNA here.
Analytical Technologies
All of our gene editing projects are tested at multiple stages to ensure high levels of engineering success. Nuclease activity is verified, clones are screened using appropriate assays such as junction PCR, and modifications are confirmed by sequencing. For more challenging or demanding projects, Cell Design Studio uses additional modalities to enrich for correctly modified cells or confirm the desired modification.
Droplet Digital PCR (ddPCR)
Droplet Digital PCR is used to characterize the expected frequency of homology-directed repair (HDR) in targeted cells and develop a screening strategy based on this expected frequency. In some cell lines, HDR occurs at a low frequency. In such cases we have utilized ddPCR to screen very large cell pools (105) in order to identify unique clones having the desired mutation. Digital PCR is also used to accurately and expediently measure target gene copy number, including in polyploid cell lines. In the final step of cell line engineering, ddPCR has proven useful to ensure clonality of cell lines.
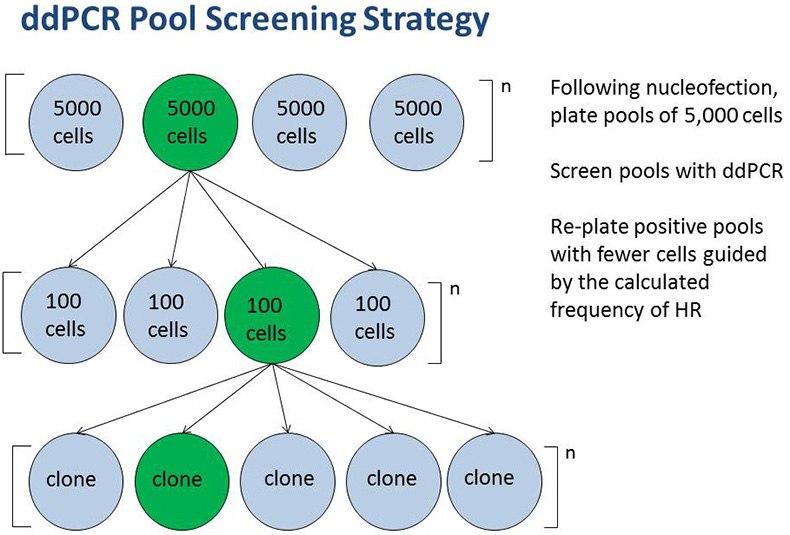
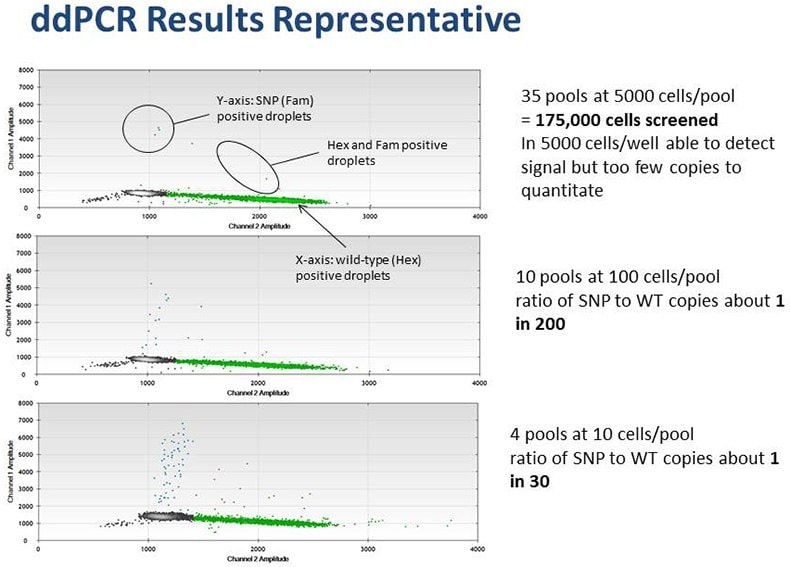
Example of a ddPCR-based pool screening strategy to isolate rare clones.
Flow Cytometry and FACS
Cell Design Studio often uses fluorescent proteins to facilitate the cell engineering workflow. As with ddPCR, flow cytometry can be used to characterize the expected frequency of HDR by a donor construct. Furthermore, we frequently utilize FACS to facilitate both initial bulk sorting and single cell cloning of engineered cells. Using FACS for these steps avoids the negative effects sometimes brought on during selection by antibiotics.
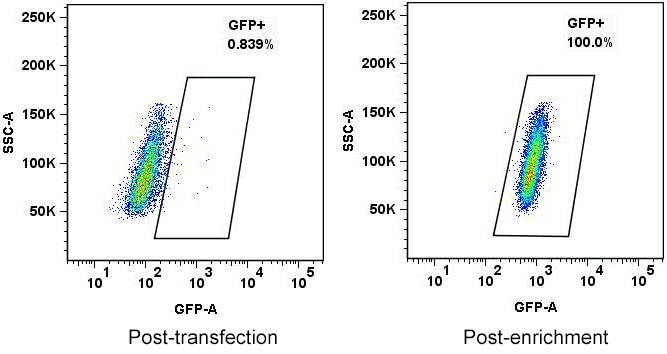
HEPG2 cells transfected with ZFNs to tag an allele with GFP. The left panel shows the cell population immediately following transfection; the right panel shows cells that were flow sorted to enrich for GFP+ cells following transfection.
如要继续阅读,请登录或创建帐户。
暂无帐户?