Chronobacter spp. Classic and New Detection Methods
Jvo Siegrist
Microbiology Focus Edition 1.2
Enterobacter sakazakii, now reclassified as a novel genus called Cronobacter, is known to be related to neonatal infections.
Cronobacter is a rod-shaped, motile and facultatively-anaerobic bacteria of the family Enterobacteriaceae. Originally Cronobacter spp. were listed as yellow-pigmented Enterobacter cloacae (Table 1). The bacteria was then later on called Enterobacter sakazakii. Taxonomic studies have determined that E. sakazakii comprises a high genetic heterogeneity and should be reclassified as a novel genus, “Cronobacter”1.
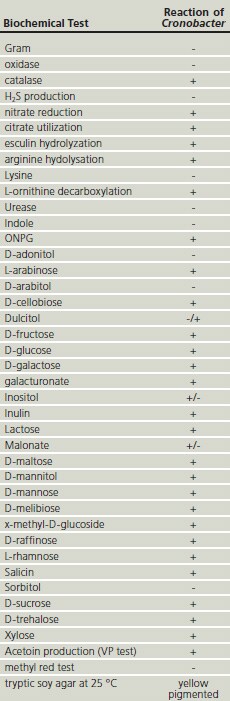
Table 1.Biochemical reactions from Cronobacter spp.

Cronobacter spp. are ubiquitous and frequently found on vegetable, meat, fermented bread, diary products and especially in baby food. Consumption of contaminated powdered infant formula milk (IFM) can result in sepsis, infant meningitis and necrotising enterocolitis. In most cases the victims are pre-term infants, low birth weight infants or immunocompromised infants in the first weeks. Mechanisms are not fully understand now but it is assumed that endotoxins are produced. The bacterium can adhere to and penetrate into various types of cells (e.g. endothelial cells). It also survives in macrophages.
As Enterobacteriaceae are suspectible to heat they do not survive most production processes and it is most likely that contamination happens after the process. The post process contamination can come from the addition of heat sensitive additives such as vitamins or other micronutirents or incorrect handling while reconstitution or storage. It has been shown, that stationary phase Cronobacter spp. are remarkably resistant to osmotic and dry stress compared with other species of the Enterobacteriacae group. It is difficult to isolate Cronobacter spp. from samples as they are most likely stressed, unevenly distributed throughout the batch and also numbers will probably be fairly low, often lower than 1 CFU per g.
For the classical microbiological tests a pre-enrichment is used to recover the stressed cells followed by a selective enrichment step.
The FDA method recommends Enterobacteriaceae enrichment (EE or Mossel) broth which is then streaked onto VRBG agar and suspect colonies are subcultured onto TSA agar where the yellow pigmented colonies are confirmed by oxidase test and other biochemical tests. See also Table 1 with all kind of biochemical reaction from Cronobacter spp. species (not only from FDA). ISO/TS 22964:2008 methods recommends buffered peptone water (BPW) as pre-enrichment medium and modified lauryl sulphate broth with vancomycin (mLST) incubated at 44 °C for the secondary selective enrichment step. The next step is then a chromgenic agar for isolation and identification (see Table 2).
There are diverse chromogenic agars available which help to save work and time to result and are more reliable than traditional media. The detection principle is based on the alpha-glucosidase possessed by Cronobacter spp. (not by most other Enterobacteriaceae) which cleaves the 5-bromo-4-chloro-3-indolyl-a-D-glucopyranoside or similar substrates. The result is a plate with e.g. blue colonies in case of Cronobacter spp. (Figures 2 and 3) but biochemical confirmation is still required.
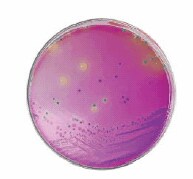
Figure 2.HiCrome™ Cronobacter spp. Agar (92324); Cronobacter spp. (blue), E. aerogenes (green) K. pneumoniae (yellow)
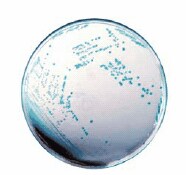
Figure 3.HiCrome™ Cronobacter spp. Agar, Modified (14703), with Cronobacter spp. colonies
Today there are also studies that not all Cronobacter spp. giving yellow pigmented colonies on tryptic soy agar and it was showed that some type of strains did not grow at 44-45 °C. Also some selective media may be too selective to recover all the Cronobacter species. There is still some work to do to improve the official methods.
A New Molecular Biology Method
Rapid detection and identification of Cronobacter species is required, since even low cell numbers have been reported to cause a disease. HybriScan®D Cronobacter spp. is a new rapid molecular test system for detection of bacteria of the genus Cronobacter in food, especially in dried infant formula milk and its production environment. It is based on the detection of rRNA by sandwich hybridisation and so no PCR is needed. It is a 96 well microplate format and the workflow is very similar to an ELISA test.
Figure 4 shows the specificity of HybriScan®- Cronobacter spp. Different cell amounts and related Enterobacteriaceae were tested within a validation study. No signals were observed using 2,3 x 108 Enterobacter cloacae cells or 7 x 108 Citrobacter freundii cells per assay, whereas clear specific signals were detectable using 2,6 x 103, 1,3 x 104, and 2,6 x 104 cells of Cronobacter species, respectively. These results demonstrate that the HybriScan® system is highly specific for Cronobacter spp.
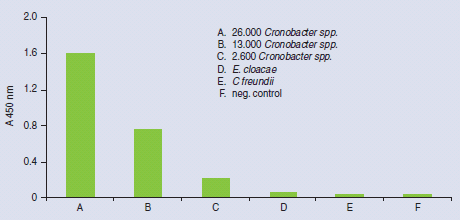
Figure 4.Specificity of HybriScan®-Cronobacter spp. Different cell numbers of Cronobacter spp. and related Enterobacteriaceae like E. cloacae and Citrobacter freundii were tested. Measurement data for HybriScan® analyses represent absorption at 450 nm.
A validation study of HybriScan®- Cronobacter spp. was performed using two different enrichment procedures: (1) single step enrichment for 24 – 26 hours at 37 °C in ESSB broth (Enterobacter sakazakii selective broth) and (2) two step enrichment starting with a pre-enrichment for 18 – 20 hours at 37 °C in buffered peptone water and followed by a selective enrichment for 24 – 26 hours at 45 °C in mLST selective broth. The results of the above mentioned validation study are presented in Figure 4.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?