Step-by-Step Protocol for Procainamide Labelled Glycan Profiling of a Monoclonal Antibody
Romana Rigger, PhD<sup>1</sup>, Pegah Jalili<sup>2</sup>, Uma Sreenivasan, PhD<sup>3</sup>, Kevin Ray, PhD<sup>2</sup>
1Buchs, Switzerland, 2St.Louis, MO, USA, 3Round Rock, TX, USA
Workflow for released N-glycan analysis
A complete workflow based on UHPLC-FLR-MS is developed to analyze the N-glycan profile. The workflow offers the following:
- Step-by-step instructions for sample preparation and analysis
- Procainamide labeling for increased detection sensitivity
- Separation using BIOshell™ Glycan HPLC column
- Low flow rate for reduced solvent consumption
- Compatibility with mass spectrometry
Characterization of Monoclonal Antibodies- Analysing the N-linked Glycan of Adalimumab by UHPLC-FLR-MS and Procainamide Labeling
Careful and thorough characterization of therapeutic mAbs is essential to ensuring drug safety and efficacy. Hence, establishing several critical quality attributes (CQAs) for each therapeutic protein, and demonstrating that production batches are within acceptable limits, is necessary for both innovator and biosimilar therapeutics. In this application note, we describe glycan analysis of the innovator mAb adalimumab (Humira) as well as a recombinant mAb of the same sequence.
Monoclonal antibodies (mAbs) are target specific and have high efficacy and few side effects. Glycosylation is one of the most common and important post translational modifications for mAbs. Glycans attached to the antibodies play an important role in the pharmacokinetics, efficacy, and safety of therapeutic adalimumab. Glycosylation involves the attachment of glycans at specific sites on a protein, most commonly at asparagine (Asn, N-linked) or serine/threonine (Ser/Thr, O-linked) amino acid residues. There are two types of glycosylation; N-linked glycosylation and O-linked glycosylation, and both types are important for protein conformation, protein activity, providing protection from proteolytic degradation, and intracellular trafficking and secretion. N-glycan moieties also play a key role in the folding, processing, and secretion of proteins from the endoplasmic reticulum (ER) and the golgi apparatus. Based on the large influence of glycosylation on protein function, an accurate study and analysis of glycans is essential. Protein glycosylation is specifically mentioned in established technical guidelines, e.g., ICH Q5E and Q6B and FDA’s published guidance for industry titled “Development of Therapeutic Protein Biosimilars”.
There are four options to approach N-glycan analysis: intact glycoproteins, glycopeptides, released glycans, or monosaccharides. This article focuses on the analysis of released N-glycans by UHPLC, combined with fluorescence (FLR) and mass spectrometric (MS) detection. The analysis of released N-glycans presented here is one of the most powerful and commonly used approaches for glycan composition analysis. Fluorescent derivatization allows relative quantification of percent abundances of glycan species by fluorescence detection. Among fluorescent derivatization molecules, procainamide offers one of the best signals, surpassing the more traditional 2-AB and 2-AA labeling. In addition, procainamide bears a tertiary amine giving it good ionization for glycan characterization by electrospray mass spectrometry. Hydrophilic interaction liquid chromatography (HILIC) is a proven technique for the separation and quantitation of glycans and has notable advantages over the other HPLC separation modes (e.g. reversed-phase, anion exchange). In this protocol, a BIOshell™ Glycan HPLC column is used to analyze adalimumab N-glycans labeled with procainamide.
Adalimumab is a recombinant human IgG1 monoclonal antibody (mAb) specific for human tumor necrosis factor (TNF). It has a molecular mass of about 150 kDa and is N-glycosylated on the Fc region.
General procedure- Sample Preparation and System Setup for Released N-linked Glycan Analysis using UHPLC-FLR-MS
Samples
Two samples are analyzed and compared using the following protocol:
- Adalimumab Reference (Humira®, from Abbvie, Inc., North Chicago, IL
- MSQC16 SILu™Lite SigmaMAb Adalimumab Monoclonal Antibody (equivalent, recombinant protein from Merck)
Reagent preparation
Buffers and Enzymes
- 8 M Guanidine HCl, prepared in water
- Dissolve 7.6 g guanidine HCl in water and bring the final volume to 10 mL
- 0.2 mL/sample (152.85 mg guanidine HCl/sample) is required
- 50 mM Ammonium bicarbonate (ABC buffer), prepared in water
- Dissolve 395 mg ABC in 100 mL water
- 2 mL/sample (7.91 mg ABC/sample) is required
- 1 unit/µL PNGase F
- Dissolve in water; may be aliquoted and stored for 6 months or longer at -20 °C
System suitability reagents
The workflow, including glycan release, labelling, and SPE clean-up, is also performed on an IgG purified from human serum (hIgG) as a control handled alongside other samples.
A procainamide labelled dextran hydrolysate is used for a glucose unit (GU) ladder to assist with interpretation of sample glycans. When analyzed on the BIOshell™ Glycan HPLC Column, the GU ladder gives a characteristic profile, from monomeric glucose to approximately a 20-mer of glucose oligosaccharide. The ladder provides calibration reference points that can aid in identifying more complex glycans based upon retention characteristics.
- hIgG (I4506) for system suitability
hIgG, IgG purified from human serum, (200 μg) is processed as a system suitability control.- Prepare a 10 µg/µL solution in 8 M guanidine HCl and aliquot in 200 µg portions
- IgG is handled identically to the samples
- Store any unused portions at -20°C for later use
- Dextran hydrolysate, procainamide labelled, for HPLC-FLR-MS system suitability GU ladder
- Dextran hydrolysate is solubilized in 25% 75 mM ammonium formate / 75% acetonitrile (v/v), and procainamide labelled according to 2.4
N-Glycan release
Prior to the analysis, samples are reconstituted to a concentration of 1 mg/mL in water and 200 µg of each protein is used for N-glycan analysis.
Denaturation
- Set heating block to 50 °C
- Start with at least 100 µL protein at neutral pH
- Add 200 µL 8 M guanidine HCl solution and quickly vortex to mix
- Incubate at 50 °C for 30 min to denature (shaking optional)
- Bring the temperature of the sample to RT
Buffer exchange to 50 mM ABC buffer
- Transfer sample to a 30 kDa spin filter
- Centrifuge at 14,000 x g for 15 min
- Add 400 µL ABC buffer
- Centrifuge at 14,000 x g for 30 min
- Repeat the previous two steps once more, making sure all the solution has passed through the filter
- Discard the flow through and place filters in new collection tubes
Enzymatic release of glycans
- Set up heating block to 37 °C
- Add 50 µL of 50 mM ABC buffer to each filter unit
- Add 4 µL of 1 UN/µL PNGase F to each filter unit
- Cap and seal centrifuge device with parafilm
- Incubate at 37 °C for 14-20 hours with shaking at 300 rpm
Note: The digestion time can be decreased to only 30 min using PNGase Fast (EMS0001-1kt) which produces a comparable result for most antibodies.
Recovery of glycans
- Centrifuge at 1,000 x g for 10 sec to collect lid condensate
- Add 40 µL ABC buffer
- Centrifuge at 14,000 x g for 5 min
- Add 100 µL ABC buffer
- Centrifuge at 14,000 x g for 5 min
- Repeat previous two steps once more
- Transfer glycans from collection tube to 0.6 mL microcentrifuge tubes for labelling
- Evaporate the solvent by vacuum centrifugation
Procainamide labelling
Dried samples are labelled with procainamide in a one-pot reductive amination solution, purified by normal-phase SPE[CM3], and the resulting labelled product is then dried again. Glycans are solubilized in 50 µL of 25% 75 mM ammonium formate/75% acetonitrile (v/v) prior to UPLC-FLR-MS.
Note: All preparation and labeling must be performed in a fume hood except for weighing reagents. Prepare the incubation block by moving to the fume hood and set the temperature to 65 °C.
Procainamide labelling reagent
- Weigh at least 1.8 mg sodium cyanoborohydride (NaBH3CN) per labelling reaction in a tube
- Tare a microcentrifuge tube
- Transfer NaBH3CN to the tube in the fume hood; a pencil eraser-head volume is usually sufficient
- Cap the tube and blow off any dust with N2 gas in the fume hood
- Weigh the tube
- Weigh at least 2.033 times more procainamide hydrochloride, by mass, than NaBH3CN in a separate tube
- Prepare 9.1 µL of a 70% dimethyl sulfoxide (DMSO)/30% acetic acid (AcOH) (v/v) solution per mg procainamide
- Solubilize the procainamide with the 70% DMSO/30% AcOH (v/v) solution
- Ensure the solution is homogenous by vortexing
- Add 18.5 µL of solubilized procainamide per mg of NaBH3CN
Note: NaBH3CN will not completely solubilize; As exposure to strong acid releases cyanide gas, this step especially warrants working in the fume hood - Add 5 µL water per mg of NaBH3CN to completely dissolve NaBH3CN
- Cap and mix by vortexing in the fume hood to fully solubilize NaBH3CN
Procainamide labelling of glycans
- Add 40 µL labelling reagent per sample to reaction solution
- Vortex for 1 min and briefly centrifuge
- Place capped tube in incubator block and incubate at 65 °C for 3 hours
Note:Cover with foil to limit condensation on the lid and keep dark
SPE clean up
- Prepare glycans for clean up by adding 200 µL of 70% ACN in water solution to the labelled glycans
- Prepare Discovery Glycan SPE tubes and vacuum manifold
- Place centrifuge tube under cartridge for waste collection
- Wash with 1 mL water, with minimum pressure gradient by vacuum manifold
- Equilibrate with 1 mL 99% ACN in water, with minimum pressure gradient by vacuum manifold
- Stop flow when meniscus completely enters top frit
- Loading the samples
- Place microcentrifuge tube under cartridge for breakthrough collection
- Add full sample volume to cartridge bed
- Pass sample through bed by gravity
- When meniscus completely enters top frit, add 500 µL of 99% ACN
- Pass volume through by gravity, collecting in same tube
- Stop flow when meniscus completely enters top frit
- Place centrifuge tube under cartridge for waste collection
- Add breakthrough + 99% ACN to bed
- Pass volume through bed by gravity
- Stop flow when meniscus completely enters top frit
- Wash
- Add 1 mL of 99% ACN, and pass the solution through cartridge with minimum pressure gradient by vacuum manifold
- Repeat the above step four more times
- Elute
- Place new microcentrifuge tube under cartridge for purified glycan collection
- Add 200 µL of 20% ACN to bed
- Pass volume through bed by gravity
- When meniscus completely enters top frit, repeat the previous two steps once more
- After the collection drip has stopped, apply medium vacuum manifold pressure to evacuate all liquid from SPE to the collection tube (Total volume ~400 µL)
- Evaporate the solvent by vacuum centrifugation
- Dried labeled glycans can be stored at -20 °C for at least 6 months
UHPLC-FLR-MS
Solubilize glycans
- Dissolve the dry glycans in 50 µL of 75% ACN / 25% of 75 mM ammonium formate (v/v) pH 4.4 (adjusted with formic acid) and vortex for 2 min
- Centrifuge at 16,000 x g for 2 min
- Transfer 40 µL to autosampler vials
UHPLC-FLR-MS parameters
The instrumentation is configured so that total flow from LC passes first through the fluorescence detector. The flow is then split 1:1 so that half goes to the mass spectrometer and half to waste. In this way, a blockage in the line to the electrospray emitter will not damage the flow cell of the fluorescence detector.
Sample analysis
- Run 1-2 blanks at the start of sample list
- Analyse dextran ladder and hIgG samples first
- After every 5 samples, run a blank followed by the dextran GU ladder to update GU values
- Run a blank after samples and before column flush
- At completion of queue, flush column with water for 30 minutes followed by 80% ACN/20% water for 30 minutes and store
Data analysis
LC peaks are identified by their level of residue composition (Table 1) from the calculated glucose unit (GU) of their elution. GU levels are determined for each LC feature’s retention time by interpolation to a 5th-order polynomial standard curve of the dextran hydrolysate ladder chromatogram (Figure 1). Glycan assignment is done by comparing the GU values to a custom database of glycan GU values for the BIOshell™ column. The dextran ladder is analysed via UHPLC FLR-MS after every fifth sample to correct for any retention time shifts. For relative quantification, fluorescence peak areas are normalized to the sum of all identified glycan fluorescence peak areas. The limit of quantification (LOQ) is defined as 0.5% of the most abundant peak area. This parameter allows for compositions less than 0.5% of the total peak area because composition is normalized to the sum of all peak areas. The peak area is calculated using Xcalibur Qual Browser. For general sample analysis, the Thermo Xcalibur Qual Browser software retrieves and records the base peak intensity for each sample.
Released N-linked Glycan Analysis Results
System suitability results
The procainamide-labeled dextran GU ladder and control acquisitions demonstrated that the UHPLC-FLR system and column were suitable to resolve and identify labelled oligosaccharides, as per the four system suitability requirements shown in Table 2. First the workflow, including glycan release, labelling, and SPE clean-up, was tested with hIgG and found to be acceptable, with 14 common hIgG glycans detected. A minimum of 10 hIgG glycan identifications are required. The second system suitability requirement was also met; peaks G1F (1,6) and G1F (1,3) were observed to be partially resolved in the fluorescence chromatogram (Figure 1). Although resolution between the G1F (1,6) and G1F (1,3) peaks was not complete, they could be visually differentiated upon magnification. The third system suitability requirement was also met by the hIgG control. The relative abundances of a representative subset of glycans from this sample gave an R2 correlation of 0.99 with the historical glycan profile. Finally, the slope created during analysis of the hIgG control was observed to be 0.99, which is within the slope requirement of 1.00 ± 0.07.
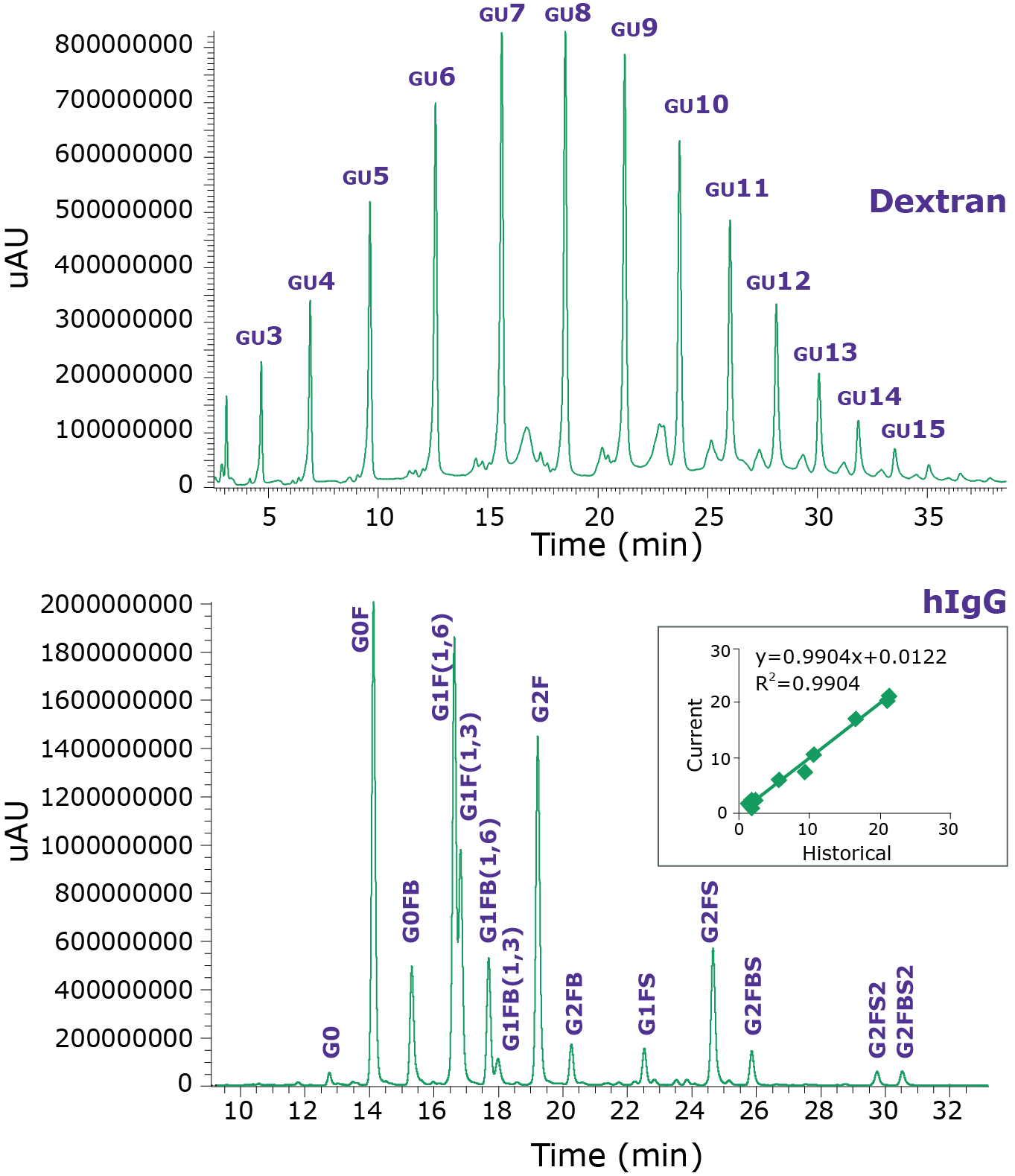
Figure 1.Fluorescence chromatograms of procainamide-labeled dextran GU ladder and hIgG control. Top pane: annotations indicate the number of glucose units (GU) in each dextran hydrolysate-procainamide feature. Each feature of the control hIgG in the bottom pane was similarly identified. The inset shows the correlation of the glycan features’ relative compositions with the historical values of previous hIgG data.
Adalimumab sample results
Sixteen glycan features for adalimumab samples were quantified. Table 3 contains glycan compositions for all the samples. Figure 2 illustrates the fluorescence chromatograms for each sample type of adalimumab. Overall, the N-glycan profiles are broadly similar, but some differences exist in the observed relative compositions of some components. Table 4 illustrates the structures of the observed glycans. The area under the peaks of glycans in the TIC chromatogram are summed and MS spectra were created to confirm the mass of glycans. Figure 3 shows the typical examples of MS spectra.
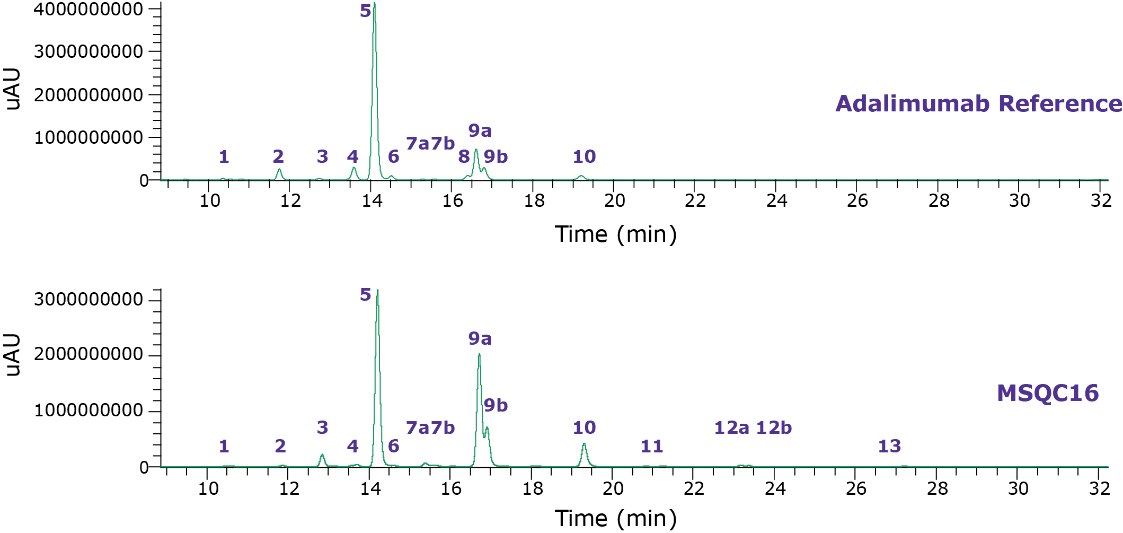
Figure 2.Fluorescence chromatograms of adalimumab samples; Reference (top) and MSQC16 (bottom). Numbered features are tabulated in Table 3.
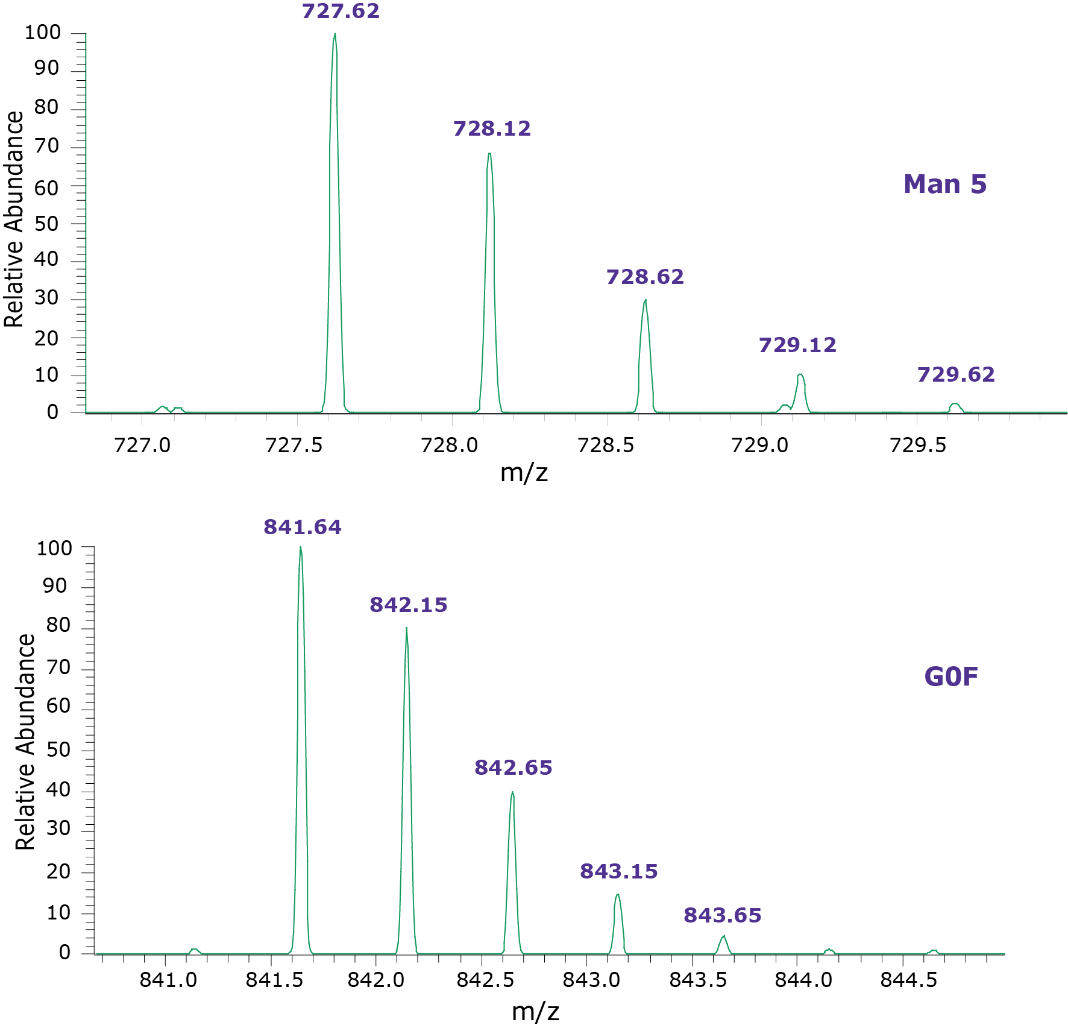
Figure 3.Typical examples of MS spectra observed for peak 4 (Man 5) and 5 (G0F) from Figure 2.
Conclusion
A complete UHPLC-FLR-MS workflow has been developed to simplify the analysis of N-linked glycans. This workflow offers the following:
- MS and Fluorescence compatibility
- System suitability testing using human IgG
- Rapid and reproducible N-Glycan release. The protocol involves multiple wash steps and an overnight digestion of the denatured mAb
- Procainamide labelling ensuring high fluorescence intensity and electrospray ionization efficiency while showing comparable chromatographic separation in comparison with other fluorescence labelling systems
- BIOshell™ HPLC column for UHPLC-FLR-MS analysis: suitable for analysis of protein-linked glycans with typical mobile phases used for hydrophilic interaction liquid chromatography (HILIC)
- Complete listing of all reagents, consumables, and related products.
A total of 16 glycan features were quantified for the mAb adalimumab. The glycan profile, including the qualitative and quantitative aspects, is comparable to the results found by other analytical laboratories. 1, 2
Characterizing and monitoring the glycosylation pattern of therapeutic mAbs is required by regulatory authorities to ensure efficacy and safety of these drugs.
This detailed protocol can be used for the analysis of N-linked glycans of mAbs and for complex and heterogenous glycoproteins.
References
To continue reading please sign in or create an account.
Don't Have An Account?