PCR-based Assay Regulations and Validation
A Technical Guide to PCR Technologies
On This Page
Regulatory Landscape Overview
While many PCR assays are developed for research applications there are further considerations for those that are being developed to become diagnostic assays or to be performed in support of: Biologics License Application (BLA), New Drug Application (NDA), Premarket Approval (PMA), 510(K) or other regulatory filing. These will need to be validated and performed in accordance with the regulated environment. The regulated environment requires quality assurance group oversight and that work is carried out in compliance with quality systems under the appropriate regulations. This is dependent on the purpose for which the PCR-based assay is used.
There are multiple regulations that are governed by several international and country-specific agencies, covering different products and industries. For example, qPCR based assays are the most sensitive for detection of residual DNA that is considered to be an impurity and remains after manufacturing of biopharmaceuticals or vaccines that are produced using live systems such as microbial or mammalian cell cultures. The requirements for assay sensitivity can vary from product to product due to differences in the dose and the size of the host genome. However, varying amounts of residual DNA may be suggested as acceptable by different regulatory agencies, e.g., less than 100 pg/dose by U.S. Food and Drug Administration (FDA)1 but less than 10 ng/dose by the World Health Organization (WHO)2 and European Agency for the Evaluation of Medicinal Products (EMEA)3. At the same time, regulations are changing with the progress of the industry, analysis of more data and efforts for harmonization. Examples of the key agencies are presented in Table 12.1.
Examples of the most common regulations that are governing PCR based assays include Good Laboratory Practice (GLP), Good Manufacturing Practice (GMP), or Clinical Laboratory Improvement Amendments (CLIA) regulations.
GLP regulations are often mistaken for general good laboratory practices, for example those addressing the use of protective clothing such as gloves and lab coats or the use of standard operating procedures. In reality, GLP is a set of regulations addressing performance of “the non-clinical safety testing of test items contained in pharmaceutical products, pesticide products, cosmetic products, veterinary drugs as well as food additives, feed additives, and industrial chemicals”4. GLP regulations direct how these laboratory studies are planned, performed, monitored, recorded, reported and archived.
World Health Organization defines GMP as the aspect of quality assurance that ensures that medicinal products are consistently produced and controlled to the quality standards appropriate to their intended use and as required by the product specification. GMP defines quality measures for both production and quality control and defines general measures to ensure that processes necessary for production and testing are clearly defined, validated, reviewed and documented, and that the personnel, premises and materials are suitable for the production of pharmaceuticals and biological products, including vaccines. U.S. Food and Drug Administration (FDA) refer to cGMP as “systems that assure proper design, monitoring and control of manufacturing processes and facilities. This includes establishing strong quality management systems, obtaining appropriate quality raw materials, establishing robust operating procedures, detecting and investigating product quality deviations and maintaining reliable testing laboratories”5.
As defined by CDC, the Clinical Laboratory Improvement Amendments of 1988 (CLIA) regulations include federal standards applicable to all U.S. facilities or sites that test human specimens for health assessment or to diagnose, prevent, or treat disease. CDC, in partnership with Centers for Medicare & Medicaid Services (CMS) and FDA, supports the CLIA program and clinical laboratory quality. The CMS regulates all laboratory testing (except research). The objective of the CLIA program is to ensure high quality laboratory testing.
Rapid technology development coincides with quick changes in instrumentation and analysis software. That creates an additional opportunity and pressure for pursuing the newest developments. Researchers supporting different phases in the drug or development process (discovery versus manufacturing, for example) will have different needs and sets of requirements for reagents, instruments and software based on the regulatory environment that also will need to be considered during experimental design. While researchers doing discovery in a non-regulated environment can work with “research use only” reagents, equipment and software and adopt new advances in technology, translational scientists working with assays supporting clinical trials are required to work within regulatory constraints. This difference is also apparent in the case of diagnostic assay development and the significant burden of validation requirements can be a limiting factor in the choice of platforms and reagents.
Assay Validation
In addition to the evaluation of the assays, as discussed in PCR/qPCR/dPCR Assay Design, assays that are used in regulated environments must be taken through analytical validation and documentation in accordance with ICH guidelines that are recommended for adoption by the regulatory bodies of the European Union, Japan and USA6 for registration of pharmaceuticals for human use. Diagnostic assays need to be validated in compliance with CLIA/College of American Pathologists (CAP) requirements. Generally, these regulations have similar requirements for assay validation but also each has specific differences that need to be considered during assay validation protocol design.
ICH guidelines Q2(R1)6 address typical validation characteristics which should be considered and they have been applied to bioanalytical method validations, including PCR-based methods, in EMEA guidelines issued in 20117. The guideline focuses on the validation of the bioanalytical methods generating quantitative concentration data used for pharmacokinetic and toxicokinetic parameter determinations. Guidance and criteria are given on the application of these validated methods in the routine analysis of samples from animal and human studies.
The main characteristics of a bioanalytical method that are essential to ensure the acceptability of the performance and the reliability of analytical results are: Selectivity (specificity), lower limit of quantification, the response function and calibration range (calibration curve performance—linearity and range), accuracy, precision, matrix effects, stability of the analyte(s) in the biological matrix and stability of the analyte(s) and of the internal standard in the stock and working solutions and in extracts under the entire period of storage and processing conditions.
The guidelines address criteria for acceptance during validation and analytical runs, as well as key components for reports. CLIA and CAP requirements for diagnostic assay validation have slight differences. CAP requires parameters 1–6 to be evaluated for both FDA cleared and Laboratory Developed Tests (LDT) assays, while CLIA allows for FDA cleared tests to be evaluated according to parameters 1–4 only. At the same time, CLIA has an additional “other” category that is defined as “any other performance characteristic required for test performance”.
Regulatory Parameters
1. Reportable Range
2. Precision
3. Accuracy
4. Reference Range
5. Analytical Sensitivity
6. Analytical Specificity
7. Other
There is often confusion regarding terminology, particularly pertaining to assay validation versus qualification.
Prior to validation, each assay should undergo a set of assay optimization experiments that will establish assay performance. These data will be used for establishing the assay validation protocol. Well-conducted optimization experiments will greatly reduce the chance of assay failure during validation.
Assay Validation: Full set of evaluation experiments (for the parameters listed above) performed in accordance with a validation protocol to demonstrate and document performance of an assay and set specifications and criteria of acceptance for expected assay performance during testing.
Assay Qualification: Assay qualification is a reduced set of evaluation experiments that mostly focus on demonstrating that an established assay will produce expected results for the intended role (for example, specific type of samples or conditions).
Approved Tests
PCR-based techniques are considered by industry and regulatory agencies as one of the “gold standard” technologies that are used for validation of results obtained by different assays and technologies when applicable, during 510(k) and PMA applications. For example, for validation of array-based genotyping assays or platforms, polymorphism, genotype call validation can be performed by PCR amplification of the target region followed by sequencing and/or by detection with qPCR probe or primer-specific assays. There are multiple regulatory guidelines that address specific assay types.
The majority of PCR-based tests that are being approved for diagnostics are focusing on detection of viral agents and mutations associated with genes involved in cancer pathways.
During the initial nine months of 2013, four out of 22 FDA approved medical devices and tests were PCR-based assays (18%), all of them were supporting personalized medicine and three out of four were companion diagnostic tests (Table 12.2)8-11. By identifying patients who have particular mutations, such as EGFR exon 19 deletion or exon 21 (L858R) substitution in non-small-cell lung carcinoma (NSCLC) cells, the test results are used to stratify the subpopulation of patients who will have a higher chance of responding to GILOTRIF®, a drug that is directed towards blocking abnormal function of the mutant EGFR protein.
Laboratory tests that are applied to identifying which particular strain of Hepatitis C virus (HCV) the patient is infected with9 are an important factor in ensuring better treatment outcomes for patients with chronic HCV infections.
Evaluated together with other clinical data tests support a personalized choice of appropriate therapy since different HCV genotypes respond differently to available drugs.
Despite fast technological advances, regulatory approval process for new assays may take several years and currently PCR-based applications are taking a more prominent role in assays that are passing the approval process and replacing classical methods, such as culture, not only in the diagnostic arena but also for release testing of biologics.
The MycoTOOL® PCR Mycoplasma Detection Kit-based test was approved by FDA on November 1st, 2012. It is the first commercially available Mycoplasma PCR test approved by FDA that can replace conventional and time-consuming mycoplasma detection assays (culture methods as well as indicator cell culture method) for the testing of biologics and biopharmaceuticals12. This test can be performed in a few hours and replaces up to 28 days processing, thus significantly improving time for lot release of biopharmaceuticals and vaccines.
Conclusions
Polymerase Chain Reaction (PCR) based methods are powerful techniques that can provide scientifically sound data, even within the budget constraints that researchers are currently experiencing. This manual provides an introduction to PCR, qPCR, RT-PCR and digital PCR with an overview of technical options and related applications, alongside guidance for troubleshooting PCR-based data. This handbook has been designed to support scientists who have different degrees of familiarity with PCR-based methods and serve as an introduction to the technology, a reference guide and a tool for daily use for researchers in academic, as well as industrial and diagnostic laboratories.
Different PCR-based methods are used by the scientific community and the choice of technique adopted is based on
application, which range from a classical end-point PCR when a fast and simple “yes” or “no” answer is needed, to digital PCR to detect rare mutations. As summarized in Introduction and Historical Timelines, PCRbased technology is mature and well established; significant knowledge has been accumulated over the last four decades and thousands of scientific papers are being published every month that use PCR-based techniques. That number has continued to grow linearly over the last 20 years, as can be seen in Figure 12.1.
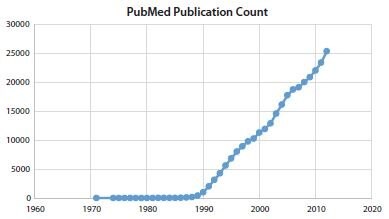
Figure 12.1.The number of records published per year (1971–2012) resulting from a search of NCBI PubMed database13 when the key word “PCR” was used.
As with every technology, PCR-based methods have limitations. This manual provides information, strategies, comparisons of different detection systems and analysis methods (see Data Analysis) to assist researchers in the appropriate evaluation of these limitations and to design an optimal experiment. As described in Quantitative PCR and Digital PCR Detection Methods, there are different strengths for each qPCR detection chemistry method and these need to be considered during the experimental design to balance sensitivity, specificity, cost of reagents and resources and time available for assay optimization. The same consideration should be applied when evaluating experimental design using individual vs. multiplex assays.
This manual also provides options for appropriate experimental design and workflow. All steps in the process, from the quality of the sample to data analysis method, as well as a choice of normalizing genes for determination of relative RNA expression, must be considered and addressed during experimental design to reduce variability. A primary concern when working with PCR is to control for cross contamination to avoid false positive results because these techniques are highly sensitive.
There are different approaches to the laboratory set up that can assist in addressing that concern, including use of proper laboratory practices with a focus on cleaning, physical segregation with dedicated equipment and gowns and a unidirectional workflow. Proper cleaning must be performed with nucleic acid destroying agents such as a simple 10% bleach solution and UV irradiation (not to be replaced with common hospital disinfectants like Amphyl® that only have antimicrobial properties). Segregation can be a simple separation into one place to store reagents and set up the master mix for PCR reactions with addition of template and controls (“pre-PCR area”) and a second place for re-amplification and data generation (“post-PCR area”). This approach is commonly adopted in a research laboratory setting. At the opposite extreme, separation would include a combination of dedicated, segregated differentially pressurized rooms with separate air handling systems, dedicated personnel, equipment, gowns and a unidirectional workflow. These separation methods are more often adopted in production and diagnostic environments14. It can be challenging to ensure segregation in a small research laboratory where space is limited and equipment cannot be dedicated. In this case, the focus must be on stringent cleaning procedures before, after and between the steps, use of filtered pipet tips, bench top workstations, and maximum possible physical separation. These measures can produce successful results. However, additional controls need to be incorporated into experiments to demonstrate lack of cross-contamination. An example workflow is presented in Figure 12.2.
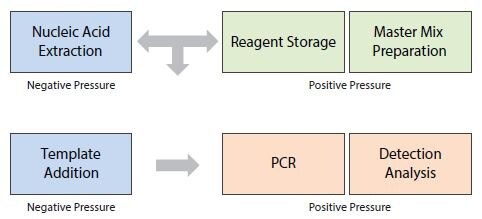
Figure 12.2.Example of a Laboratory Workflow: Division of functional areas and the workflow direction in the PCR assay process.
The importance of the quality of template and choice of appropriate QC assays is discussed in Sample Purification and Quality Assessment and Reverse Transcription. Comparing RNA samples that have undergone different degrees of degradation can lead to erroneous biological conclusions about RNA expression signatures. Similarly, different samples may have different concentrations of inhibitors that will impact RT or PCR efficiency and may produce false-negative results or inaccurate ratios of expression. For example, there is a vast diversity of sample types that are being interrogated with PCR-based techniques; multiple environmental samples tested to evaluate biodiversity or to detect pathogens, different human and animal tissues and bodily fluids to evaluate RNA expression signatures or DNA vaccines. Each of these can have a plethora of unique inhibitors that may impact different assays in the same sample to a variable degree.
Different biological questions can be answered by utilizing multiple workflows, reagents, instruments and applications, as discussed throughout this guide. The choice of an optimal technique, reagent, method of analysis, types and number of replicates and controls will be dependent on multiple factors and constraints of individual projects as well as the researcher’s experience and personal preferences. However, as discussed in Quantitative PCR and Digital PCR, significant emphasis must be placed on maintaining compliance with MIQE guidelines15 through all phases of the experiment to assure scientific integrity of the data and validity of biologic conclusions that will withstand the test of time and follow up experiments16.
Next-generation Sequencing
All PCR-based techniques are applied to targeting known sequences, as discussed in PCR/qPCR/dPCR Assay Design. While sequencing methods can generate and detect “unknown” sequence, PCRbased amplification is an important step in the generation of template for massively parallel sequencing (MPS). Quantitative PCR approaches are used for QC and optimization of DNA libraries and this is a key step in the generation of high-quality data. This approach is highly sensitive and requires a very small amount of material, compared to other methods. In addition, qPCR evaluation assures a high efficiency and thus cost effectiveness of sequencing data. PCR is also a corner stone for template generation for popular technologies used for resequencing of DNA and RNA. These applications are supported by design tools that allow multiplexing of over 6,000 primers per pool and 10 ng of starting material, making interrogation of limited human samples such as FFPE material feasible.
However, together with the trend of reduction of cost of MPS and increase in depth of coverage per sample, nextgeneration sequencing instruments are rapidly evolving. RNASeq applications are approaching not only sequence determination, but also quantification of transcripts and new platforms are approaching sequencing of single molecules without amplification. When the technology reaches this point, it may well replace PCR-based diagnostics (rapid point of care detection of infectious pathogens or known cancer mutations such as in EGFR gene for example). However, that is still in the future. PCR-based applications continue as a significant work engine supporting research, development and diagnostics.
Additional Technical Support
This guide is provided to researchers to address the most common PCR related questions. In addition to the troubleshooting guide, the scientific group and technical support team is also available to assist with further support, troubleshooting or, in cases such as when follow up experiments need to be conducted in a regulated environment, outsourcing to reference laboratories. Our goal is to enable our customers to succeed.
References
To continue reading please sign in or create an account.
Don't Have An Account?