CRISPR on the Brain? Standardized Functional Genomics Tools Advance Neuroscience
Deciphering the genetics of neuroscience has always been challenging. Heterogeneous tissue microenvironments, complex genetic interactions and phenotypes, lack of model systems that accurately mimic the human brain transcriptome – not to mention the blood-brain barrier – make genetic perturbation analysis difficult in neural tissues1,2. In the last decade, the advance of functional genomics tools has begun to reveal some of the relationships between genes and neurological phenotypes.
In this article, we describe:
- The journey toward global access to standardized functional genomics
- How genetic modulation using RNAi has revealed key genes in neural pathways
- How genome editing has enabled the development of more predictive model systems with which to interrogate the functional genomics of the nervous system
Functional Genomics: The Need for Standardization
Upon the sequencing of the human genome, some believed that the connection of all genotypes to phenotypes could not be far behind. Within a few years, functional genomics tools, such as RNAi reagents and zinc finger nucleases were developed to modulate gene expression with the hopes of connecting the altered state to a phenotype. However, early uses of these tools yielded variable results, depending on their precise sequence, bioavailability, processing efficiency, knockdown efficiency, undocumented off-target effects, and other parameters3. Genome-wide studies, with the additional variability introduced by pooling strategies and library size/depth, also lacked reproducibility.
Driven by the need for standardized reagents, The RNAi Consortium was born in 2005, centered at the Broad Institute. This collaboration, which included Sigma-Aldrich (now MilliporeSigma, the Life Sciences business of Merck KGaA, Darmstadt, Germany), consisted of 11 organizations. Over ten years, TRC developed 3 generations of RNAi reagent libraries, culminating in the 2015 launch of a 350,000 clone, mouse+ human shRNA library. The former TRC, now known as the Broad Institute’s Genetic Perturbation Platform, has made its shRNA libraries available through www.sigmaaldrich.com/rnai, which is the only source for all glycerol stocks and all lentiviral formats for these libraries.
Driving Global Access to Standardized Genome Editing
In 2007, together with Sangamo Biosciences, we launched the first commercially available genome editing reagent, our CompoZr® Zinc Finger Nucleases. This award-winning gene editing technology enabled us to co-publish a paper reporting the first transgenic rat developed using a genome editing tool4, and to develop novel cell lines with target genes endogenously tagged with fluorescent reporters.
In 2013, one year after the first report of CRISPR/Cas9 function, we were the first to release a globally available, commercial CRISPR reagent portfolio. After we licensed key CRISPR patents from the Broad Institute, our CRISPR portfolio has rapidly expanded to include alternative sgRNA delivery formats, pooled sgRNA libraries, and most recently, the Sanger Whole Genome CRISPR Arrayed Libraries. While the majority of genome editing researchers are using the tools to study cultured cells (according to an informal survey of our customers), a sizable and growing number are editing genomes to generate transgenic animals, enabling heretofore impossible interrogations of the complex brain.
How Standardized Functional Genomics Tools Have Advanced Neuroscience Research
Neural phenotypes, as well as neurological and psychiatric disorders, are rarely caused by a single gene. It is no wonder that forward genetics using perturbations of individual genes is an inefficient method for discovering the basis of complex behaviors. Using RNAi to modulate combinations of genes has yielded more insight into neural pathways. Such RNAi studies appear to be most robust when they use standardized reagents with well-characterized specificity, knockdown efficiency, common controls, library depth, and sequence accuracy.
Neuroscience Research Highlight: RNAi Exposes Trio of Trafficking Proteins
The neurological symptoms of pseudorabies virus infection are notorious: pigs show ataxia, cattle suffer itchy skin, canines howl. The virus exerts these effects by infecting neurons and traveling up axons toward the cell body. How does the virus travel? Researchers at Princeton University used RNAi reagents from MilliporeSigma to silence genes that they suspected might be involved in axonal trafficking of this virus5. Their data showed that three proteins -- a dynein regulator, a vesicle transport regulator, and an intermediate filament – were primarily responsible for viral transport (Figure 1).

Figure 1.Compared to control siRNA, siRNAs specific for three axonal trafficking proteins reduced virus transport when transfected into neurons prior to viral infection. Adapted from Koyuncu OO, Perlman DH, Enquist LW. Cell Host Microbe. 2013 Jan 16;13(1):54-66.
Neuroscience Research Highlight: Female, CRISPR-Generated Mutant Mice Show Anorexia-Like Behaviors
Eating disorders are perhaps the epitome of complex neuropsychiatric phenotypes. In 2013, the histone deacetylase 4 (HDAC4) gene was linked with an increased risk for developing eating disorders. Researchers at the University of Iowa recently reported that they used the CRISPR/Cas9 system to generate a transgenic mouse bearing the specific A778T HDAC4 mutation that was previously linked to eating disorders in humans6 The authors reported that, compared to wild type mice, the HDAC4 mutant mice did not try as hard to obtain more food when they were hungry (measured by the number of “nosepokes” the mice performed to try to access a food pellet; Figure 2). This failure to respond to hunger cues is reminiscent of anorexia nervosa. The mice also exhibited obsessive-compulsive tendencies and abnormal responses to social interaction. Surprisingly, these altered behaviors were only observed in female mice.
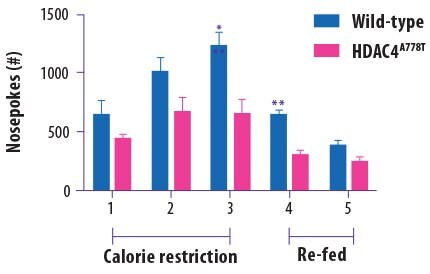
Figure 2.Female wild-type mice poked their noses to access food pellets more times than female HDAC4 mutant mice, suggesting that this genetic mutation is linked to a blunted response to hunger cues. Adapted from Lutter M et al. Biological Psychiatry. 2016 Oct 13.
Neuroscience Research Highlight: Genome Editing Used to Create “Alzheimer’s Neurons”
Familial Alzheimer’s Disease (fAD) has been linked to specific mutations, but the mechanism of action by which those mutations cause disease are not fully elucidated. Researchers at the University of California, San Diego, used genome editing to generate mutant induced pluripotent stem cell (iPSC)-derived neurons that bear mutations in presenilin and amyloid precursor protein (APP)7. Compared to wild-type neurons, the mutant neurons showed abnormalities in the Rab11-mediated axon transport pathway, leading to altered localization of APP and low-density lipoproteins (Figure 3).
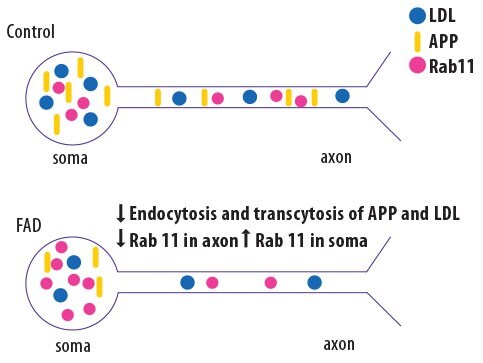
Figure 3.Schematic showing the results of analyzing gene-edited “Alzheimer’s neurons.” IPSC-derived neurons bearing fAD mutations (bottom), a decrease in Rab11 levels in the axons results in reduced trafficking of APP and low density lipoprotein (LDL) compared to wild-type neurons (top). Adapted from Woodruff et al. Cell Rep. 2016 Oct 11;17(3):759-773.
What’s Next? Continued Development and Validation of Functional Genomics Tools
To be a true partner to researchers who wish to take advantage of functional genomics tools, life science suppliers need to be users as well as manufacturers of these tools. In parallel, they must maintain close collaborations with the leading research laboratories, together driving innovations in genomic editing.
As we continue to develop reliable, reproducible tools based on the technologies we have licensed from the Broad Institute, we also build our Core Partnership Program, a community of core laboratories who often collaborate with our genomic editing experts to develop model systems, create cell lines, and identify new genes of interest.
Visit our functional genomics learning center and get your brain hooked on discovery. We’re just getting started.
References
To continue reading please sign in or create an account.
Don't Have An Account?