Overview of Cancer Biology
During the last twenty years our understanding of the biology of cancer has increased dramatically. It is now widely recognized that cancer results from a series of genetic alterations causing a loss of normal growth controls, resulting in unregulated growth, lack of differentiation, apoptosis, genomic instability, and metastasis. Cancer knows no boundaries and can develop in any tissue of any organ at any age. However, one of the hallmarks of tumor development is a long latent period with no obvious clinical evidence of disease. Epidemiological studies have demonstrated that the incidence of cancers increases exponentially with age. These studies suggest that three to seven rate-limiting mutations are required for full development of cancer. Indeed, this stepwise progression of cancer is supported by clinical human data, as well as in animal carcinogenesis models (Figure 1).
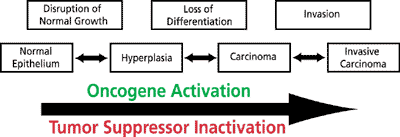
Figure 1. Carcinoma development and invasion.
The upper row represents disturbances in growth, differentiation, and tissue integrity that lead to the phenotypes that characterize the different stages of cancer, shown in the lower row. Multiple genetic alterations underlie cancer development, including oncogene activation and tumor suppressor loss of function.
The genetic and biochemical, defects that are required to transform a normal cell into a cancerous cell mass with abnormal growth and an ability to invade adjacent tissue remains largely unresolved. At the molecular level, in order to gain the proliferative advantage required for tumorigenesis, cells must have alterations in the genes that are responsible for cell cycle progression and growth. These mutations alter the levels or function of the proteins encoded by growth-regulating genes, effectively altering cell division. The two major categories of genes mutated in cancer are oncogenes and tumor suppressors.
The majority of oncogenes were originally identified as altered forms of proto-oncogenes acquired from the RNA genome of retroviruses (v-onc). In the early 1900s Rous demonstrated that transplantable sarcomas in chickens could be induced by a cell-free agent. This cell-free agent was identified as a retrovirus that had transduced part of a normal cellular gene, src (sarcoma). The virally transduced gene (v-src) was mutated compared with its cellular counterpart (c-src), rendering it constitutively active. Over the past two decades, many different retroviruses have been identified and shown to induce tumors, each containing a different oncogene. The mechanism of oncogene activation by a provirus is shown in Figure 2. Table 1 includes a list of identified oncogenes, their genetic lesion, and neoplasia type.
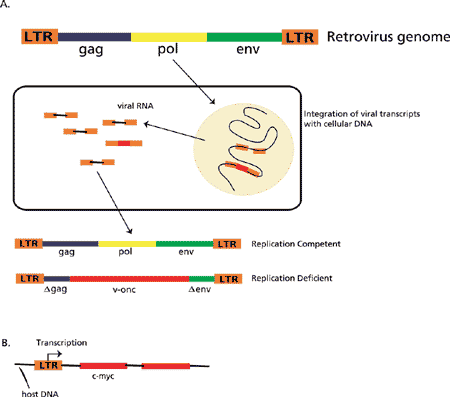
Figure 2. Mechanism of activation of a host gene by insertion of a provirus.
A. The retroviral genome is composed of a gag region that encodes the internal structural proteins of the virion, the pol region that encodes the virion RNA-dependent DNA polymerase (reverse transcriptase; RT), and the env region that encodes the proteins found on the surface of the virion envelope. The LTR region is the long terminal repeat that appears at each end of the integrated linear DNA. Within the LTR are specific DNA sequences that define the initiator site for RNA transcription and a poly(A) site on the 3’ end where the viral polyadenylation occurs. In addition, there are transcription enhancer sequences that result in the production of high levels of transcripts. The elements within the LTR provide all of the necessary functions for eukaryotic transcription to occur and for the provirus to express genomic viral RNA. The virion RT converts the retroviral genome into DNA that is subsequently inserted into the host cell chromosomes and replicated by means of the host cell transcription enzymes. Viral genetic material that has become integrated into host germ cell chromosomes becomes inheritable, and it is estimated that 3% of the human genome is derived from ancient retroviral genetic material. If the inserted viral genome remains intact, it is replication competent in that it can replicate intact virus particles. However, if, for example, pol is replaced with other genetic material, such as a viral oncogene (v-onc), an intact virus cannot be replicated and the genome is replication deficient.
B. Insertion of a proviral LTR upstream of the first coding exon of an oncogene (e.g., c-myc) results in a transcript that no longer contains c-myc regulatory sequences. The c-myc gene becomes constitutively expressed, and cellular c-Myc levels increase.
Besides viral activation, oncogenes can also be activated via chromosomal translocations, overexpression, and gene amplification. Cellular oncogenes have been found in multiple copies in many different tumor types and transformed cell lines. The mechanism by which gene amplification bestows a selective advantage to the cancer cells is by overexpression of genes contained within the amplicon. All amplified oncogenes express high levels of RNA and protein and do not appear to have had gene rearrangement occur.
The c-Myc gene was the first oncogene shown to be amplified. Many other oncogenes have been shown to be frequently amplified in human cancers including c-myb, the Myc family, the Ras family, Mdm2, and the erbB and FGFR families. The presence of multiple copies of oncogenes in cancer cells has been associated with poor prognosis. N-Myc, which was first identified in neuroblastoma, is present in multiple copies in approximately 40% of neuroblastomas and its amplification correlates with late stages of the disease. Amplification of c-Myc and N-Myc is also observed in small cell lung cancer and appears to be associated with the more advanced and malignant stages of the tumor. In addition, amplification is observed with Ras genes, which encode G proteins involved in the signaling cascades of tyrosine kinase receptors. Amplification and overexpression of Ras leads to constitutive signal transduction, such as that observed with continual activation of growth factor receptors, such as epidermal growth factor receptor (EGFR) or platelet-derived growth factor receptor (PDGFR). This activation leads to continuous cell proliferation and ultimately cancer.
As stated above, a third mechanism of oncogene deregulation is through chromosomal translocations. It has been demonstrated that several oncogenes are located at or near chromosomal translocation breakpoints. In most cases, structural gene rearrangements that juxtapose two different chromosomal regions are believed to contain genes that are dominant for transformation, such as oncogenes. However, chromosomal deletions are thought to be the sites of tumor suppressor genes. Recurring translocations were first identified in cytogenetic analyses of leukemias and myelodysplasias. These chromosomal anomalies of leukemias and lymphomas are generally somatically-acquired alterations. One example is the translocation of c-Abl and Bcr, t(9;22) (Figure 3) resulting in a chimeric protein, Bcr-Abl.
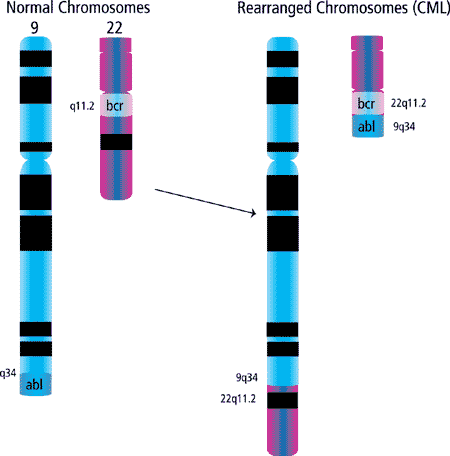
Figure 3. Generation of the Philadelphia chromosome observed in >95% of chronic myelogenous leukemias (CML).
The Bcr-Abl fusion protein stimulates unregulated growth and has been shown to be involved in the pathogenesis of chronic myeloid leukemia (CML) and a subset of acute lymphoblastic leukemia (ALL). Although many translocations give rise to new chimeric proteins, some result in altered regulation of the oncogene. A prime example is in Burkitt's lymphoma, where the c-Myc locus on chromosome 8 is translocated to a position distal to the immunoglobulin heavy chain locus on chromosome 14 (t(8;14)). Whereas the expression of c-Myc is normally tightly regulated, in cells with this translocation c-Myc is constitutively expressed. Many other translocation breakpoints and candidate oncogenes have been identified.
On the other hand are the tumor suppressor genes, which normally block the development of malignancies by encoding for proteins that suppress tumor initiation and growth. From his epidemiologic studies of retinoblastoma, Knudson proposed his "two-hit hypothesis" to explain mechanisms involved in inactivating tumor suppressor genes during tumorigenesis. The first mutation could be germ line (inherited) or somatic, while the second mutation was almost always somatic
This hypothesis illustrates the mechanisms through which inherited and somatic mutations might collaborate in tumorigenesis. Over twenty tumor suppressor genes have been identified through various approaches (Table 2). Based on comparisons between cancer tissue and corresponding normal tissues, the identification of regions with a high frequency of genomic deletion detected by allelic loss or loss of heterozygosity (LOH) has been used to localize genes that predispose to cancer (tumor suppressor genes).
Tumor suppressor proteins have been implicated in a wide array of cellular processes, including cell cycle control, cell-cell adhesion, differentiation, apoptosis, and maintaining genomic stability. One of the best studied is the tumor suppressor gene retinoblastoma (Rb), which encodes for the protein pRB. The pRB protein plays a role in cell cycle progression through its ability to silence the expression of E2F target genes, such as those required to drive cells through S phase. Many sporadic cancers display an inactivating mutation in the Rb gene, while in other cancers the pRb pathway is inactivated as a result of mutations to other mediators in the pathway. Another important regulatory tumor suppressor protein is p53, which functions to prevent replication of damaged DNA in normal cells and promotes apoptosis in cells with damaged DNA. p53 is the most highly mutated protein in all human cancers. Inactive or mutated p53 allows cells with damaged DNA to survive and proliferate. These mutations are then passed to daughter cells, conferring a high probability of neoplasm.
Telomeres are nucleoprotein complexes at the ends of eukaryotic chromosomes consisting of many double stranded TTAGGG repeats. Two of the main functions of telomeres are to mark the end of a linear chromosome as distinct from a broken DNA end, and to facilitate chromosome replication. It is well known that telomeres play a critical role in the maintenance of chromosomal integrity, a fact that has since been confirmed in diverse model systems. However, telomere lengths are markedly different, even among mammals. For example, human telomeres are 5 to 15 kb long while those of inbred mice may exceed 60 kb. Telomeres are maintained by a reverse transcriptase, the ribonucleoprotein telomerase, which is composed of a RNA subunit, telomerase RNA component (TERC), and a protein catalytic subunit, telomerase reverse transcriptase (TERT), whose expression is tightly regulated. Telomerase expression is low or completely absent in most human somatic tissues, with its expression in the adult restricted to activated lymphocytes, germ cells, and tissue stem cells. The vast majority of cells that exhibit telomere dysfunction undergo senescence or apoptosis, and different lines of evidence have shown that these processes are important barriers to tumorigenesis. Telomerase activation is one of the most commonly observed features of cancer, as it is seen in greater than 80% of all human tumors. Over-expression of telomerase enhances the transformation of human cells in vitro and tumorigenesis is blocked by telomere shortening in animal models of cancer. Together, these data suggest that telomere-induced checkpoint activation is a major in vivo tumor suppressor mechanism.
Environmental factors may also play key roles in tumorigenesis. Viruses that have been linked with human malignancies include human papillomaviruses (cervical carcinoma), cytomegalovirus (Kaposi's sarcoma), Epstein-Barr virus (Burkitt's lymphoma and nasopharyngeal carcinoma), and hepatitis B virus (hepatocellular carcinoma). Retroviruses have been linked to T-cell lymphomas (human T-cell lymphotrophic virus; HTLV-1), which have a predilection for skin and bone involvement, hypercalcemia, and a leukemic phase. The mechanism of HTLV-1 neoplastic transformation is integration of the provirus (double-stranded DNA copy of the viral RNA genome) into the cellular genome. Parasites, such as Schistosoma haematobium, have been linked to bladder cancer, and Opisthorchis sinensis has been linked to carcinoma of the pancreas and bile ducts. Overexposure to ultraviolet radiation is a cause of skin cancer (i.e. basal and squamous cell carcinoma, melanoma, and especially in xeroderma pigmentosum). Ionizing radiation is also carcinogenic; survivors of the atomic bombs dropped in Hiroshima and Nagasaki have a higher-than-expected incidence of leukemia and several other cancers. Similarly, the use of ionizing radiation to treat nonmalignant diseases (thymic or adenoid enlargement and ankylosing spondylitis) also increases the incidence of cancer in these individuals.
While the genetic basis of tumorigenesis may vary between cancer types, the steps, both cellular and molecular, required for metastasis are basically the same. In normal tissue, homeostasis is maintained between epithelial cells and their microenvironment, such as vascular endothelial cells, fibroblasts, immune cells, and the extracellular matrix (ECM). In the cancerous state, these interactions become deregulated. As a tumor begins, nutrients are provided by direct diffusion from the circulation. Local tissue invasion can result in pressure on normal tissues, which can lead to inflammation, or the tumor may produce substances (e.g. collagenase, gelatinase, stromelysin) that lead to enzymatic destruction of tissues. Subsequently, synthesis of tumor angiogenesis factors cause formation of an independent vascular supply to the tumor. From inception, a tumor may begin to shed cells into the circulation. Indeed, from animal studies, it is estimated that a 1-cm3 tumor can shed greater than 1 million cells per 24-hour period into the venous circulation. Most circulating cancer cells die as a result of intravascular trauma, thus the greater period of time that a tumor cell spends in the circulation, the greater the chance of its death. The probability of a circulating tumor cell becoming a metastatic tumor is estimated at less than one in a million.
The process of metastasis involves interplay between ECM degradation, proteolysis, cell adhesion, cell migration, angiogenesis, and tissue homing. Metastases develop when cancer cells adhere to the vascular endothelium and penetrate into surrounding tissues, surviving and spawning independent tumors at distant tissue sites.
Once at the new site, tumor growth resumes, disrupting normal tissue and organ function, thus continuing the process. Studies suggest that metastasis is not a random event and that the primary tumor may actually regulate the growth of metastatic tumors. For example, in renal cell carcinoma, the rate of tumor growth is often similar in the primary and metastatic nodules. In some cases removal of the primary tumor has resulted in rapid growth of the metastases.
With advances in our understanding of the signaling pathways regulating the processes of tumorigenesis and metastasis, we hope to one day find novel and better-targeted regimen for both the prevention and treatment of cancer.
References
About the Author
Dr. Troy Baudino is currently an assistant professor in the Department of Cell and Developmental Biology and Anatomy at the University of South Carolina School of Medicine in Columbia, SC. Dr. Baudino's research focuses on Myc oncoproteins and their roles in development, angiogenesis, and cancer. He received his B.S. in Biology from Bradley University in May 1993. Following a two-year stint in the pharmaceutical industry he decided to attend graduate school. He went on to receive a Ph.D. in Cellular and Molecular Biology from St. Louis University in May 1999 where his mentor was Dr. Paul MacDonald. Dr. Baudino continued his training as a post-doctoral fellow in the laboratory of Dr. John Cleveland at St. Jude Children’s Research Hospital in Memphis, TN.
To continue reading please sign in or create an account.
Don't Have An Account?