Genomic Analysis of Formalin-Fixed Paraffin Embedded (FFPE) Tissues through the use of Whole Genome Amplification (WGA)
Ernest Mueller
The past is a source of knowledge, and the future is a source of hope. S. Ambrose (1936-2002)
Preserved samples from medical, forensic, museum and other archival collections represent a rich source of study material, much of it meticulously collected, characterized and preserved through many decades of work by experts in the field. While the majority of this material was originally collected and preserved with morphological studies in mind, the advent of molecular genetic techniques have caused many scientist to eye archival collections as a precious source of biological data.
There are many preservation methods that have been applied to tissues, including alcohol preservation, formalin treatment, freezing and sequestration in waxes and other materials. Molecular diagnostic research groups began working with formalin-fixed paraffin embedded (FFPE) tissues in the late 1980s and early 1990s .1,2 Their results showed some early successes. Unfortunately, many of the preservation methods used in the past, of which formalin treatment has been the most popular, have been found to degrade DNA and RNA. Common techniques of extraction and molecular analysis that worked well with fresh and frozen samples were found to work poorly on samples stored in unbuffered formalin or FFPE preserved samples. This so-called “formalin problem” appears to be worse for samples that have been stored over longer periods. It has been hypothesized that while the immediate adducts created by formalin are reversible, the slower reactions are irreversible and leave nucleic acids unsuitable for PCR, hybridization and other molecular biology methods.3,4 Strand breaks or formaldehyde lesions in the DNA are equally able to ruin amplification.
Researchers have attempted to characterize the effect of long-term formaldehyde preservation on DNA. While the conditions of fixation are myriad and the possible products complex4, past researchers have shown that the ability to analyze FFPE derived material in applications such as microarrays is loosely related to the average length of extracted DNA.5 More recent and apparently more correlative diagnostics for template quality have been developed which use the ability to PCR amplify from the sample. 6,19,20 One assay, in particular, has worked well in our hands as the best predictor of a sample’s potential for successful amplification.7 The assay employs multiplex PCR to amplify a number of differently sized loci within the human genome; loss of the higher molecular weight bands indicates that the DNA has degraded past the point where whole genome amplification will be successful. This test allows scientists who are interested in amplifying a limited sample of unknown quality to perform some polymerase-mediated amplification before attempting WGA.
Any WGA method to be applied to formalin-fixed DNA must be able to amplify relatively short lengths of nucleic acid stretches, ~1kb or less. Four WGA methods have been described and are in common use – they include linker adapter PCR (LAP)8-10, primer extension preamplification PCR (PEP)11, degenerate oligonucleotide PCR (DOP)12,13 and multiple strand displacement (MSD).14,15 While all have been successful in varying degrees when used with FFPE DNA, MSD has suffered in that it relies on a highly processive polymerase and high quality DNA chains to create long amplicons (>10kb). Because FFPE DNA is often comprised of short pieces of DNA with amplification terminating lesions, this type of input DNA has been amplified with best representation by using LAP and DOP WGA .10,13,16
GenomePlex® uses a modified DOP amplification to effect whole genome amplification, and the method has been successfully applied to formalin-fixed samples.16 There has been some confusion as to the mechanism of this system, which was first described in 2004 by Pruett et al.17 GenomePlex uses a mild fragmentation step to randomly break up long DNA, which facilitates subsequent steps. Priming is accomplished using an oligo with known 5’ ends but degenerate 3’- ends, and the multicopy DNA sample is reproduced in a series of overlapping single stranded DNAs. These single stranded products are subsequently primed with the same oligo to create double stranded DNA with fixed 5’- ends. The resulting library is amplified using PCR, Figure 1 .
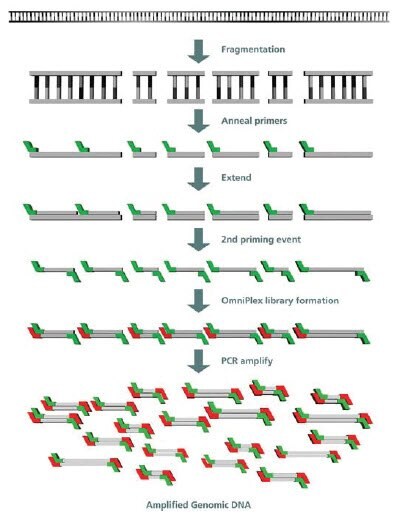
Figure 1. GenomePlex Mechanism
The GenomePlex hybrid method has several advantages over the previously described WGA methods. It does not require template with long, amplifiable regions, as does MSD. The method does not rely on highly efficient digestion and ligation as does LAP amplification. Finally, GenomePlex has improved on PEP and DOP amplification with the design of improved primers, and does not suffer from reduced amplification due to competing primer interactions.
There are limits to all WGA techniques, and tissue fixation by formaldehyde is a highly varied procedure that affords extracted DNA of vastly different qualities. As related above, MSD requires relatively long stretches of amplifiable DNA to create a representative product. LAP, in contrast, relies on both a highly efficient restriction digest and complete ligation in order to deliver representative amplification. In contrast, GenomePlex primers and cycling parameters have been optimized to prime, on average, twice for every ~1 kb stretch of genomic DNA. Therefore, when starting with genomic DNA fragments that have been severely degraded (below 1 kb) GenomePlex will amplify at less than optimal efficiency, giving rise to some loss of representation of DNAs in this size range. Genomic DNA fragments less than ~400 bp will not amplify at all using GenomePlex. (18). Highly fragmented DNA (<150 bp) will, when subjected to the GenomePlex methodology, self prime to create product with very low representation as compared to the original sample.
Amplification limitations are not the only obstacle in analyzing formalin treated samples. Studies have shown that formalin treatment weakens DNA, making it more susceptible to shearing when subjected to column based bind and elute purification.4 The most recent GenomePlex offering, (Cat. No. WGA2+C4482), designed for fresh, frozen or fixed tissues, includes reagents to effect a very gentle extraction. The lysis uses a combination of proteinase K and lytic detergent, and draws from the experience gained in the development of the Extract-N-Amp™ reagent line. Efficient, gentle lysis and a robust amplification should give the greatest possible chance for the researcher to amplify their tissue-derived DNA.
In conclusion, WGA from fixed tissues is a very challenging, but rich source of potential genetic information. Because GenomePlex initially generates a library of amplifiable template molecules by relying upon a simple degenerate primer extension, it alone is poised to amplify DNAs that are highly fragmented and/or modified. The DOP amplification mechanism and the gentle lysis included in the GenomePlex Tissue WGA kit make it the best commercial choice for these difficult substrates.
Extract-N-Amp is a trademark of Merck KGaA, Darmstadt, Germany and/or its affiliates.
GenomePlex is a registered trademark of Rubicon Genomics, Inc.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?