Human Microbiome Research Workflow: An Overview of the Role in Human Health and Disease
- What factors alter the human microbiome?
- Microbiome Sequencing: 16s rRNA Sequencing
- Microbiome identification workflow and microbiome reagents
- Microbiome metabolites and short chain fatty acids
- Microbiome metabolite analysis and microbiome reagents
- Microbiome library production, bioinformatics, and data analysis
- Microbiome Materials and Reagents
How does the microbiome affect health?
Previously, the study of human microbiology was based on identification, isolation and culture of single microbes, such as bacteria, fungi and viruses. Scientists collected these organisms from patients with acute or chronic infections and designed therapies or drugs to fight these pathogens. This approach has been extremely successful in the treatment of human disease; however, it is important to note that this approach has also led to antibiotic-resistant microorganisms. Recent studies investigating the human microbiota suggest great promise for a new approach in extending human health and well-being.
What does the Microbiome Mean?
The human microbiome consists of the combined genomes of trillions of microbes in the human body, including both symbiotic and pathogenic microbes along with their genes, proteins, and metabolites1. Importantly, an imbalance of pathogenic microbes has the ability to cause life-threatening disease and illness. However, the symbiotic microbes in a healthy individual can control the environment to keep the pathogenic microbes from increasing in numbers. In some instances, when the microbiota balance is disrupted, symbiotic microbes decrease in numbers while the pathogenic microbes have the opportunity to develop and grow. It is under these circumstances when disease and illness may develop. Scientists are currently investigating the correlation between the presence of certain microbes with health and disease. The microbiome has been linked to many ailments, such as depression, anxiety, immune system disorders and obesity. It is not completely understood if the presence of certain microbes causes a disease or if a person with a type of disease has this type of microbiome because of the disease. To facilitate a researcher’s microbiome workflow, we offer a growing list of microbiome reagents, including enzymes, immune system activators, microbiome standards, antibodies, DNA-free reagents, antibiotics and staining products for microbiome research.
What factors alter the human microbiome?
Every human gut microbiota is unique, shaped early in life, and continues to change throughout our lifetime2. These microbiota remain relatively stable in adulthood but differ between individuals due to body type, exercise and diet. As a result, each person may have a specific bacterial composition that is necessary for optimal health. Many researchers are trying to understand how to change microbiome composition and the resulting impact on health and disease development3. It is unclear how microbe-containing supplements and diet modify the microbiome community, but promising research suggests that it is likely possible4. Until recently, fecal transplants have been the only effective way to modify the human microbiome. However, a new drug that modifies gut microbiome has been approved in China to treat Alzheimer’s disease, while there are some companies that offer personal microbiome analysis along with personalized nutrition plan to control diabetes and have data to show effectiveness4-5.
Microbiome sequencing: 16s rRNA sequencing
Scientists often focus on two fundamental questions when investigating the gut microbiome. The first question seeks to identify the microbial composition, while the second questions aims to understand the metabolic activity between the microbiome and the surrounding cells and tissues. In order to uncover the microbiome composition with continuously increasing sample sizes, novel culture-independent molecular and biochemical analyses, such as Next Generation Sequencing (NGS) of the 16S rRNA gene is being used at increasing scales for detection and classification of the diverse microbes present in the human body. Areas being sampled and studied include the gastrointestinal tract, skin, respiratory system, and the urogenital tract. Genomes and gene products within these regions have been widely researched and have revealed that everyone has their own microbiota and that microbiota likely plays a significant role in health and disease. This discovery could lead to personal medicine or treatments based on a patient’s individual microbiota profile.
Microbiome identification workflow and reagents
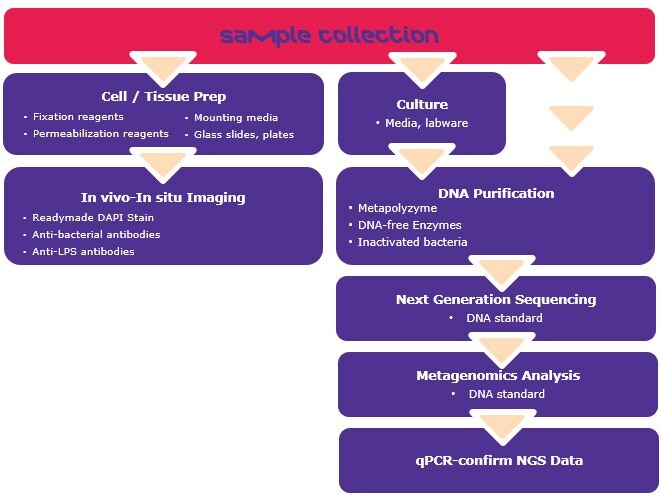
Microbiome identification workflow and reagents
Advancements in sequencing technologies have created cost-effective methods to determine the microbial composition of a sample. However, each analysis step may comprise numerous technical variables and potentially introduce variability in the acquired data. These steps include methods for sample collection without contamination, preservation and storage, sample preparation, DNA extraction and NGS sequencing6. Standardization using individual microbial DNA and inactivated bacterial standards are a major focus as to avoid bias and increase reproducibility.
Gut microbiome metabolomics and proteomics research
In addition to understanding the microbial composition, researchers are also highly interested in better understanding what the microbes are doing at the molecular level and how their presence and signaling molecules impact their surroundings. Answering these questions requires studying the relationships between the microbes themselves as well as the relationship of the microbes within the host. Understanding the interactions of both the microbiome and host genetics may provide additional insight on disease diagnosis, treatment, and prevention7-8 .
Microbiome metabolites and short chain fatty acids
In the human gut, bacteria breakdown food material and produce a byproduct which we refer to as metabolites. Microbiome researchers are interested in what these metabolites are, how they affect the microbiome composition, and how they affect the host. Short Chain Fatty Acids (SCFAs), fatty acids containing less than six carbon atoms, are an example of a bacterial metabolite that is under investigation. Interestingly, there is some evidence to suggest that this metabolite may influence gut health and may cause diseases such as Inflammatory Bowel Disease (IBD)9-10.
Researchers are using metabolomic and proteomic techniques to measure the effects of microbe and host interactions. Scientists can extract protein and metabolites from a host sample and perform mass spectral analyses, including GC-MS and LC-MS to gain relationship insight. Scientists are also beginning to find new ways to visualize these interactions by using in vivo 3D models and organoids as models to further understand the microbe and host interactions. In this type of research, effector molecules, such as metabolites, antibiotics, immune system activators, and active compounds, can be added to the in vitro host/microbe model and the effects can be studied via metabolomic, proteomic or transcriptomic methods. Bacteria-specific antibodies and FISH probes are also being used for imaging microbe-host interactions and can assist in the identification of the bacteria that are present and how they may influence the host cell.
Microbiome function analysis workflows and reagents:
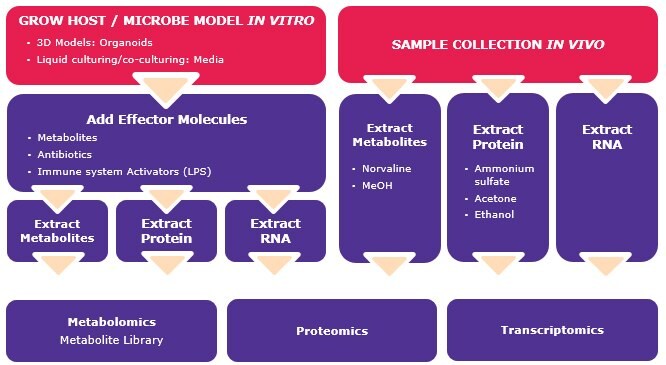
Microbiome function analysis workflows and reagents
The importance of standardized sample collection and preservation
A concern for microbiome researchers is the ability to compare data between experiments and with other research labs. It has been recognized that data collected without proper standardization is difficult to interpret. Thorough selection of experimental methods and standardizations are crucial to prevent potential bias in each step of sample collection and preservation11.
The first step in establishing a microbiome analysis pipeline is often sample collection. It is important that the sample composition remains unchanged and is as close to the original microbe composition as possible. Samples, such as mucus and urine, are easier to collect and store; while others, such as stool, blood or amniotic fluid are more challenging12. If these types of samples are not handled correctly, then the composition of the sample could change and generate bias in downstream analyses. For example, if a biological sample sits at room temperature or warmer for an extended amount of time this would promote the replication of bacteria that would benefit from this condition while others may die13. In other words, results from testing this sample would not represent the actual microbial composition that were present in the sample at the time it was collected. Standardization and controls are necessary to be able to detect if there has been changes in the sample. For example, storing samples as soon as possible at -80 °C, fixation with EtOH and addition of DNase inhibitors may increase stability of the sample’s original microbial composition.
Sample processing: preventing and monitoring DNA changes using DNA-free reagents and controls
Sample processing is a crucial step in any metagenomics project and standardization is necessary throughout the entire process. An important reason to use standards during sample processing is to monitor for DNA loss. The DNA extracted should be representative of all original microbes present in the sample. These microbes may vary in physical properties such as cell wall characteristics, gram-negative or positive. Therefore, methods to make sure sufficient amounts of high-quality nucleic acids are obtained for subsequent library production and sequencing are required. Processing requires specific protocols for each sample type, and various robust methods for DNA extraction are available14. Physical separation and isolation of cells from the samples is also important to maximize DNA yield or avoid coextraction of enzymatic inhibitors, such as humic acids, that might interfere with subsequent processing15. Enzyme mixes, such as Metapolyzme (MAC4L), a multilytic enzyme mix, can be used to ensure the digestion of difficult microbes and isolation of total DNA for metagenomics studies.
Microbiome library production
Library production for some sequencing technologies requires high nanograms or micrograms amounts of DNA, hence amplification of starting material might be required. As with any amplification method, there are potential problems associated with reagent contaminations, chimera formation and sequence bias in the amplification, and their impact will depend on the amount and type of starting material and the required number of amplification rounds to produce sufficient amounts of nucleic acids. These issues can have significant impact on subsequent metagenomic community analysis16. It is important to minimize risk of contamination by using DNA-free reagents and appropriate controls to monitor for potential bias throughout the process. Examples of microbial DNA-free reagents include lytic enzymes, such as Mutanolysin, Lysostaphin and examples for DNA extraction controls include inactivated Escherichia coli and Inactivated Proteus vulgaris.
Bioinformatics, data analysis and the knowledge to analyze data within each lab
During the last decade, sequencing tools have gradually shifted from conventional Sanger sequencing technology to next-generation sequencing (NGS), allowing for the identification of complex microbial samples. The most widely used NGS method for the taxonomic and phylogenetic evaluation of bacterial community composition relies on 16S rRNA gene amplicon analysis6. The 16S rRNA gene is approximately 1500 bp in size, consisting of nine variable regions separated by conserved regions which vary between different bacteria. Standardization of NGS is important for these methods to also avoid bias and increase reproducibility and consistency between labs. Example of DNA standards that can be used with NGS, as well as metagenomics analysis, include microbial DNA standard from E.coli and Enterococcus faecalis.
NGS is used for determining bacterial composition in a sample based on DNA fragment detection from the total DNA isolate. Predominantly for whole metagenomics sequencing, shotgun sequencing that covers the entire bacterial genome provides the most complete information on the entire gene pool within the sample. However, the large amount of generated data generated using this technique requires substantial bioinformatic efforts in sequence assembly, mapping and analyses. In many studies, both clinical and environmental, sequencing of 16S rRNA gene amplicons, covering variable regions of the gene, is the method of choice for the analyses of bacterial community composition, providing cost-effectiveness, sufficient resolution and sequencing depth17-20. The analysis of this information requires extensive bioinformatics knowledge and may not be within the range of knowledge of some bench scientists. The challenge for these labs is to train or hire someone with this extensive knowledge to interpret the data, along with complications using consistent naming conventions, various databases and tools21.
In summary, as each step in the experimental pipeline introduces variation influencing the final output, there is still an unmet need for standardization of methodology, which would enable reliable and reproducible analysis of valuable human biological samples for studying gut microbiota. We offer a growing set of standards for the microbiome workflow, consisting of microbial DNA standards and inactivated bacteria, that provides researchers with the means to normalize their results through the different stages of sample processing and analysis. Additionally, we offer a growing list of DNA free reagents, such as relevant-enzymes, immune system activators, metabolomics small molecules, antibodies, antibiotics and staining products for microbiome research.
Microbiome Materials and Reagents
References
如要继续阅读,请登录或创建帐户。
暂无帐户?