Quantitative PCR Basics
A Technical Guide to PCR Technologies
Quantitative PCR (formally quantitative real-time PCR, qPCR) detection builds on the basic PCR technique and allows researchers to estimate the quantity of starting material in a sample. Since the products are detected as the reaction proceeds, qPCR has a much wider dynamic range of analysis than conventional, end-point PCR; from a single copy to around 1011 copies are detectable within a single run. Quantitative real-time PCR and subsequent amplicon detection is carried out in a closed-tube format which eliminates the need for post-PCR manipulation, such as gel electrophoresis and significantly reduces the risk of cross contamination.
The basic principles of qPCR are discussed on this page, and the mechanisms of the common detection chemistries are described in Quantitative PCR and Digital PCR Detection Methods.
Basic Principles
When performing qPCR, a fluorescent reporter dye is used as an indirect measure of the amount of nucleic acid present during each amplification cycle. The increase in fluorescent signal is directly proportional to the quantity of exponentially accumulating PCR product molecules (amplicons) produced during the repeating phases of the reaction (see Polymerase Chain Reaction). Reporter molecules are categorized as; double-stranded DNA (dsDNA) binding dyes, dyes conjugated to primers, or additional dye-conjugated oligonucleotides, referred to as probes (Quantitative PCR and Digital PCR Detection Methods).
The use of a dsDNA-binding dye, such as SYBR® Green I, represents the simplest form of detection chemistry. When free in solution or with only single-stranded DNA (ssDNA) present, SYBR Green I dye emits light at low signal intensity. As the PCR progresses and the quantity of dsDNA increases, more dye binds to the amplicons and hence, the signal intensity increases.
Alternatively, a probe (or combination of two depending on the detection chemistry) can add a level of detection specificity beyond the dsDNA-binding dye, since it binds to a specific region of the template that is located between the primers. The most commonly used probe format is the Dual-Labeled Probe (DLP; also referred to as a Hydrolysis or TaqMan® Probe). The DLP is an oligonucleotide with a 5’ fluorescent label, e.g., 6-FAM™ and a 3’ quenching molecule, such as one of the dark quenchers e.g., BHQ®1 or OQ™ (Quantitative PCR and Digital PCR Detection Methods). These probes are designed to hybridize to the template between the two primers and are used in conjunction with a DNA polymerase that has inherent 5’ to 3’ exonuclease activity. When the DLP is free in solution, the signal intensity is low because the reporter dye is in close proximity to the quencher moiety. As more of the template is produced during the reaction, more probes hybridize to the template, which in turn are cleaved by the 5’ to 3’ exonuclease activity of the advancing DNA polymerase. The fluorescent signal intensity increases as the 5’ reporter dye is released into solution. The use of probes labeled with different reporter dyes allows for the simultaneous detection and quantification of multiple targets in a single (multiplex) reaction.
A typical qPCR run consists of repeated cycles of alternating temperature incubations, Cycling Procedure 3.1. This profile is often used when dsDNA-binding dyes, Molecular Beacons, or Scorpion® Probes are the chosen detection chemistries for qPCR. Primer extension is most efficient at 72 °C because this is the optimal temperature for processivity of most DNA polymerases. At 72 °C, polymerization occurs at a rate of approximately 100 bases per second. However, there is still processivity at lower temperatures that is sufficient to amplify shorter templates. qPCR amplicons are typically shorter (<200 bases) than conventional PCR products, thus extension is often combined with annealing in a single step at 60 °C when working with Dual-Labeled Probles (Cycling Procedure 3.2).
Cycling Procedure 3.1
An example of a standard qPCR profile using dsDNA-binding dye, Molecular Beacon, or Scorpions® Probe detection.
- Initial Denaturation: The reaction temperature is increased to 95 °C and the sample is incubated for 2–10 min (the time depends on the polymerase enzyme hot start mechanism) to ensure that all all complex targets (dsDNA) are separated and are single stranded and available for amplification
- Cycling:
- Denaturation: The reaction temperature is increased to 95 °C for 10 sec to melt all dsDNA.
- Annealing: The temperature is lowered to 60 °C for 30 sec to promote primer and probe (if included) binding to the template.
- Extension: Subsequent elongation occurs at an increased temperature of 72 °C, which is optimal for TaqDNA polymerase processivity. Duration of extension will be dependent upon amplicon size (30 sec per 1 kb). The period of elongation depends upon the desired length of the amplicon and the enzyme used. Since qPCR amplicons are short, this is typically 5–30 sec.
- Denaturation: The reaction temperature is increased to 95 °C for 10 sec to melt all dsDNA.
- Repeat: Steps 1–3 are repeated, usually for 40 cycles.
Cycling Procedure 3.2
An example two-step cycling qPCR profile for DLP detection.
- Initial Denaturation: The temperature is increased to 95 °C and the reaction is incubated for 2–10 min (depending on the hot start properties of the polymerase enzyme)
- Cycling:
- Denaturation: The reaction temperature is increased to 95 °C for 10 sec to melt all dsDNA.
- Annealing and Extension: The temperature is lowered to 60 °C for 30 sec to promote primer binding to the template and subsequent elongation occurs due to sufficient activity of the DNA polymerase at this temperature.
- Repeat: Steps 1–2 are repeated, usually for 40 cycles.
The change in fluorescence over the course of the reaction is measured by a real-time PCR thermal cycler. Dedicated real-time PCR instruments combine temperature cycling with an optical unit. The optical unit provides light at a suitable wavelength to excite the fluorophore and detects the resulting emission. Many modern instruments have an affixed display screen or nearby computer monitor, which allows the reaction to be tracked as it progresses (Figure 3.1).
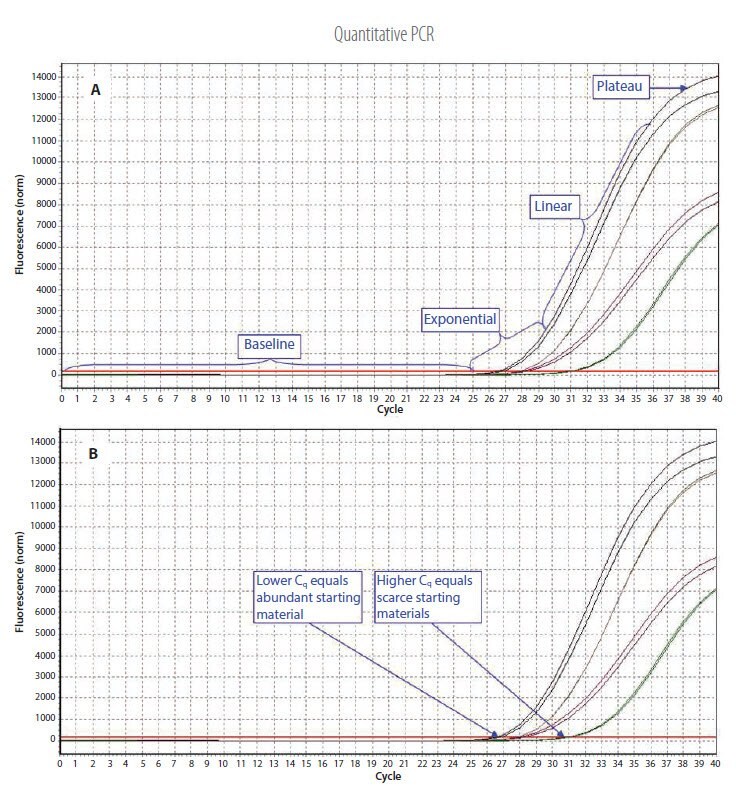
Figure 3.1.Example qPCR Assay Data. A) The different phases of the reaction. Baseline: The initial concentration of template is low; therefore, the fluorescence intensity is too low to be detected and only the background signal is evident. Exponential: After the target yield has reached the detection threshold, shown as the red threshold line, the course of the reaction can be followed through the exponential phase. Linear: As the concentration of template increases the available DNA polymerase concentration reduces and the reaction rate decreases. Plateau: The reaction is at the maximum yield. B) Individual reactions are characterized by the cycle at which fluorescence first rises above the threshold, which is referred to as the Quantification Cycle (Cq). If the starting material is abundant, amplification is observed in earlier cycles and the Cq is lower. If the starting material is scarce, amplification is observed in later cycles and the Cq is higher. This correlation between fluorescence, Cq and amount of amplified product enables quantification of the template over a wide dynamic range.
Quantification and Analysis of gDNA Targets
Quantitative real-time PCR can be readily applied to analysis of gDNA targets. Such studies may be genotyping/SNP
determination, methylation analysis, screening transgenic sequences, or monitoring of insertions and deletions.
Quantification and Analysis of mRNA Transcripts
A common application of qPCR is gene expression analysis, e.g., comparing the mRNA concentrations of a gene of interest between control and treated samples. The mRNA can be quantified via a two-step or one-step RT-qPCR process (see Reverse Transcription Quantitative Real-time PCR, for more details of RT reactions).
Two-step RT-qPCR
The reverse transcription and qPCR reactions are carried out sequentially in separate tubes. Either total RNA or, less
commonly poly(A)+ RNA, is used as the starting material and cDNA is produced by elongation from oligo-dT primers,
random primers, a blend of oligo-dT primers/random primers, or gene-specific primers using a reverse transcriptase enzyme. An aliquot of this reaction is then added to the qPCR.
One-step RT-qPCR
When the reverse transcription and qPCR reactions are combined into one tube, the experimental setup is simplified
and there is a lower chance of contamination because there is no requirement for further handling of the sample.
Quantification and Analysis of ncRNA Transcripts
Most non-coding RNAs are longer than 100 nucleotides and therefore, can be detected and quantified by RT-qPCR
with the same techniques that are used for mRNA analysis. In contrast, short non-coding RNAs, such as micro and piwi RNAs, are essentially the length of a single PCR primer. As a consequence, a technique that lengthens these short RNAs is needed before performing qPCR. Two main approaches are adopted in commercially available systems: 1) use of a stemloop RT primer and 2) poly-A tailing followed by RT with an oligo-dT adapter primer (Figure 3.2). An alternative approach developed by Casoldi et al., that is not commercially available, uses ligation of an adapter molecule to all RNA in the sample and specific primer design to differentiate between the desired targets1,2.
The stem-loop RT primer approach was developed and commercialized by Applied Biosystems (currently Life Technologies/Thermo) for subsequent TaqMan probe-based qPCR3. As illustrated in Figure 3.2A, the 5’-end of the RT primer can base-pair with a region several nucleotides from its own 3’-end to create a base-paired stem separated by an unpaired loop. All except the last few nucleotides at the 3’-end of the RT primer are universal, that is, they contain the same sequence in all miRNA primers. The last few nucleotides that extend 3’ from the stem are complementary to the 3’-end of a target miRNA. Extension of the primer along a miRNA template creates a
cDNA that can be amplified with a miRNA-specific forward primer and a universal reverse primer, the latter of which is
complementary to the 5’-end of the stem-loop RT primer.
All other commercial miRNA qPCR methods, including our MystiCq® brand, use poly-A tailing to lengthen the miRNA, as described by Shi and Chiang4. Poly(A) Polymerase (PAP), a template-independent enzyme, catalyzes the transfer of adenosine residues from ATP to the 3’-end of any RNA. As illustrated in Figure 3.2B, RT can then be performed using an oligo-dT primer. The oligo-dT primer includes an adapter sequence at its 5’-end, which enables subsequent qPCR with a forward primer that is complementary to a specific miRNA and a reverse primer that is complementary to the adapter sequence.
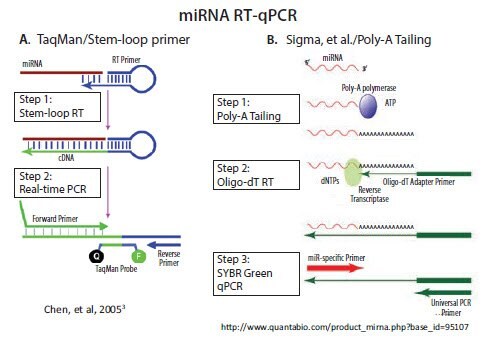
Figure 3.2.There are two commercially available methods for RT and qPCR of miRNA. A) A looped primer is used to prime the RT step after hybridization to the 3’ end of the miRNA. PCR extension through the loop results in a combined miRNA and universal sequence which is long enough for amplification B) Addition of a poly A tail to the miRNA provides a priming site for a primer that is composed of a oligo-dT tract and a universal priming sequence. Nucleic acids research by Oxford University Press. Reproduced with permission of Oxford University Press in the format reuse in a book/e-book via Copyright Clearance Center.
Both commercial approaches have advantages and disadvantages. Stem-loop priming allows 2-step RT-qPCR, whereas most poly-A tailing methods require an additional step for RT before qPCR. There are products that combine poly-A tailing and RT in a single reaction for 2-step PAP tailing/RT-qPCR. However, detection may be less sensitive with this approach (unpublished internal results). Conversely, poly-A tailing followed by RT produces a stable pool of cDNA that can be used to detect any microRNA, mRNA, or other RNA, immediately or at any time in the future. With the stem-loop/TaqMan approach, a different primer is needed for detection of each miRNA. Multiple stem-loop primers can be added to the same RT reaction5 and pools of RT primers for multiple miRNAs may be purchased from Life Technologies to perform multiplex RT. However, neither option allows for qPCR detection of miRNAs discovered at a later date, nor do they allow qPCR to detect mRNA from the same cDNA. The system described by Castoldi et al1,2 is unique in that it does provide the means to detect mRNA, precursor and fully processed miRNA molecules from the same total RNA sample.
qPCR Requirements
Instruments
Many qPCR instruments have been designed to support a specific range of applications, e.g., contrast the capability of the ABi 7900 high throughput instrument using automatic loading of 384-well plates with the Illumina produced and marketed Eco instrument that supports a single 48-well plate. Therefore, the most suitable instrument meets the needs of the research. It is desirable to select an instrument with user friendly software that performs the most desirable functions and has flexibility in terms of data output so that it can be easily manipulated in downstream statistical analysis software packages. This reduces the time required to train personnel and therefore to begin generating results. Additional features that are required include a PCR block that is absolutely uniform (an absolute maximum deviation of 1 Cq = 2-fold across 96 wells of replication) and an optical system that excites and detects emission as sensitively and as evenly as possible across a wide range of wavelengths. This allows for a wide choice of fluorophores and enables multiplexing. Other features to consider are the operating costs associated with specific consumables, e.g., if a standard microtitre plate is not used for reactions and also the convenience of loading plates/tubes that are non-standard format.
Template
Very few copies of target nucleic acid (equivalent to about 100 pg of gDNA or cDNA) are required to initiate qPCR. To minimize contamination with reaction inhibitors, the starting template amount should be kept to the minimum required to achieve accurate quantification. When the starting material is RNA, primer design and DNase I treatment will reduce signals that may be generated from gDNA contamination.
Primers
Whether using a dsDNA-binding dye or a probe-based detection chemistry, designing high-quality primers is one of the most critical pre-experimental steps in qPCR. For a detailed discussion of assay design, PCR, qPCR, dPCR Assay Design. To maximize sensitivity and specificity, any required modifications, such as Locked Nucleic Acid, should be included (qPCR Detection Chemistry).
Probe(s)
When using probes as the amplicon detection mechanism, primer dimers and nonspecific products are not detected but should be avoided because they can lower reaction efficiency. To maximize sensitivity and specificity, the appropriate probe type for the application should be selected and any required modifications, such as Locked Nucleic Acid, included (qPCR Detection Chemistry).
dNTPs
Standard PCR and qPCR master mixes contain dATP, dCTP, dGTP and dTTP. However, some mixes are available that replace dTTP with dUTP. Products from previous reactions run with dUTP will contain uracil instead of thymine. These are then susceptible to cleavage by Uracil-DNA-Glycosylase (UNG). Therefore, prior incubation of subsequent reactions with UNG prevents carryover contamination between reactions. To be effective, all PCRs in the laboratory must use dUTP.
Magnesium
Magnesium chloride (MgCl2) is necessary for reverse transcriptase, Taq DNA polymerase and Taq DNA 5’ to 3’ exonuclease activity. Optimum Mg2+ concentrations for reactions containing DLPs are usually between 3–6 mM. The majority of master mixes contain MgCl2, however, it is sometimes necessary to optimize the concentration, so an additional tube of pure MgCl2 is typically included with the master mix product (see Assay Optimization and Validation). In some cases, a reaction mix that does not contain MgCl2 may be required so that a low concentration can be used, e.g., when using Scorpions® Probe detection.
Reverse Transcriptase
A reverse transcriptase enzyme that provides high yields of cDNA, while retaining activity at high temperature, is critical to the success of RT-qPCR. Performance at high temperatures helps to ensure that regions of RNA with significant secondary structure are destabilized and accessible for hybridization and subsequent amplification. When performing one-step RTqPCR, high-temperature performance allows the use of genespecific primers with high melting temperatures (Tm), which increases reaction specificity. When performing two step protocols, it is important to ensure that the enzyme results in a linear and proportional yield of cDNA from RNA (see Reverse Transcription for details of RT evaluation).
Taq DNA Polymerase
As with selecting the most appropriate reverse transcriptase for the RT, selection of the appropriate polymerase enzyme is vital. A fundamental problem with natural Taq DNA polymerase is that the enzyme has residual activity at low temperature. Nonspecific primer binding leads to nonspecific product formation as a result of this residual polymerase activity. Antibody-blocked or chemically blocked Taq DNA polymerases (‘hot start’) help to rectify this situation by preventing enzyme activity until the high-temperature, denaturation step begins.
Buffer
Buffers or reaction master mixes, typically contain dNTPs, a Taq DNA polymerase, MgCl2 and stabilizers. SYBR Green I dye, ROX™, fluorescein and inert loading dyes may also be included (Loading Control Dyes), depending on the detection chemistry, instrument and reaction requirements. The PCR buffer components and stabilizers are typically proprietary to the manufacturer. If purchased separately, maximum flexibility is possible, since each ingredient can be optimized individually in the reaction. However, in contrast, while purchasing the ingredients together as a master mix reduces flexibility, it increases batch consistency and convenience while reducing the number of pipetting steps and hence, the chances of error and contamination.
Loading Control Dyes
Some real-time PCR thermal cyclers require a loading dye such as ROX to be included into each reaction to control for variability in the optical system and to normalize differences in signal intensity. Likewise, some thermal cyclers require an initial fluorescein signal to create a virtual background when working with SYBR Green I dye assays (which have very low background). These may be supplied in the master mix or as separate components so that the appropriate concentration can be used.
The MIQE Guidelines
One clear problem with the use of qPCR is that it is very easy to produce numbers that can then readily be interpreted as quantitative results. However, this can be equally misleading because quality control and further analysis may be required to differentiate between potential artifacts caused by inappropriate assay design, execution, or data analysis. The publication of apparently significant data without adequate quality controls has led to reports that are, at best, not biologically or clinically relevant and, at worst, are inaccurate.
Each step of the qPCR experimental process, from sample acquisition to qPCR data analysis, is vulnerable to variability6. Therefore, variability may occur during sample selection and processing, extraction of nucleic acids (can result in differences in quality which cause lack of reproducibility of qPCR target detection), assay design and optimization (contribute to differences in assay efficiency and sensitivity) and data analysis (requires some subjective interpretation and application of appropriate statistical methods). It is critical that the information provided to a scientific audience contains adequate information for the experiment to be critically assessed7, yet it is apparent that this is not always the case8. The “Minimum Information for Publication of Quantitative Real-time PCR Experiments” (MIQE) guidelines9 address these concerns. The aims of the MIQE guidelines are to increase transparency by specifying the minimum information about an assay that is required for the reader to be able to assess the technical validity of a publication and to provide guidance for design, optimization and validation of new qPCR assays.
MIQE consists of nine sections (experimental design, sample, nucleic acids, reverse transcription, target, primers and probes, assay details, PCR cycling and data analysis; visit our MIQE page to download a checklist, Table 1). All parameters relate to information that should be obtained during the process of experimental design, optimization and validation. When using commercial assays or those from the published literature, optimization and validation should still be executed. The MIQE paper includes a checklist with factors labeled Essential, which are considered indispensable for an adequate description of the qPCR assay, whereas other components are labeled Desirable, indicating that this information is useful but even without it, the assay could be replicated and the data within the publication evaluated. It is important to recognize that reporting PCR efficiency, analytical sensitivity and specificity of each individual assay is essential and should be determined by the investigator using the conditions in the laboratory. There are specific considerations when performing digital PCR (see Digital PCR) that have been addressed in a recent digital PCR MIQE publication10.
Are You MIQE Compliant?
Minimum Information for Publication of Quantitative Real-time PCR Experiments
aClinical chemical Copyright 2009 by American Association For Clinical Chemistry, Inc. Reproduced with permission of American Association For Clinical Chemistry, Inc. in the format Internet posting via Copyright Clearance center.
bAll essential information must be submitted with the manuscript, Desirable information should be submitted if available. If primers are from RTPrimerDB, information on qPCR target, oligonucleotides, protocols, and validation is available from that source.
cAssessing the absence of DNA using a no-revers transcription assay is essential when first extracting RNA. Once the sample has been validated as DNA,free, inclusion of no-revers transcription control is desirable but no longer essential.
dDisclosure of the probe sequence is highly desirable and strongly encourage. However, because all not commercial pre-designed assay vendors provides this information, it cannot be an essential requirement. Use of such assays is discouraged.
References
To continue reading please sign in or create an account.
Don't Have An Account?